Small nuclear RNA
Small nuclear ribonucleic acid (snRNA), also commonly referred to as U-RNA, is a class of small RNA molecules that are found within the nucleus of eukaryotic cells. The length of an average snRNA is approximately 150 nucleotides. They are transcribed by either RNA polymerase II or RNA polymerase III, and studies have shown that their primary function is in the processing of pre-mRNA (hnRNA) in the nucleus. They have also been shown to aid in the regulation of transcription factors (7SK RNA) or RNA polymerase II (B2 RNA), and maintaining the telomeres.
snRNA are always associated with a set of specific proteins, and the complexes are referred to as small nuclear ribonucleoproteins (snRNP) often pronounced "snurps". Each snRNP particle is composed of several Sm proteins, the snRNA component, and snRNP specific proteins. The most common snRNA components of these complexes are known, respectively, as: U1 snRNA, U2 snRNA, U4 snRNA, U5 snRNA, and U6 snRNA. Their nomenclature derives from their high uridine content.
snRNAs were discovered by accident during a gel electrophoresis experiment in 1966. An unexpected type of RNA was found in the gel and investigated. Later analysis has shown that these RNA were high in uridylate and were established in the nucleus.
A large group of snRNAs are known as small nucleolar RNAs (snoRNAs). These are small RNA molecules that play an essential role in RNA biogenesis and guide chemical modifications of ribosomal RNAs (rRNAs) and other RNA genes (tRNA and snRNAs). They are located in the nucleolus and the Cajal bodies of eukaryotic cells (the major sites of RNA synthesis), where they are called scaRNAs (small Cajal body-specific RNAs).
Classes of snRNA
snRNA are often divided into two classes based upon both common sequence features as well as associated protein factors. [1] The first, more widely studied, class is known as Sm-class RNA. Sm-class snRNA are transcribed by RNA polymerase II. The pre-snRNA are transcribed and receive a 5’ monomethylguanosine cap in the nucleus. They are then exported to the cytoplasm for further processing. In the cytoplasm, the snRNA receive 3’ trimming to form a 3’ stem-loop structure, as well as hypermethylation of the 5’ cap through trimethylguanosine. The 3’ stem structure is necessary for recognition by the survival motor neuron (SMN) complex. This complex assembles the snRNA into stable ribonucloproteins (RNPs). The 5’ cap is then required to import the snRNP back into the nucleus. The Sm-class of snRNA includes U1, U2, U4, U4atac, U5, U7, U11, and U12. All of these uridine-rich snRNA with the exception of U7 form the core of the spliceosome. Splicing, or the removal of introns, is a major aspect of post-transcriptional modification, and takes place only in the nucleus of eukaryotes. U7 snRNA has been found to function in histone pre-mRNA processing.
The second class, Lsm-class snRNAs, contain a monomethylphosphate cap and a 3' stem–loop, terminating in a stretch of uridines that form the binding site for a distinct heteroheptameric ring of Lsm proteins. Lsm-class snRNAs are transcribed by RNA polymerase III and never leave the nucleus, in contrast to Sm-class snRNA. There are only two know Lsm-class snRNAs, U6 and U6atac. [2]
snRNA in the spliceosome
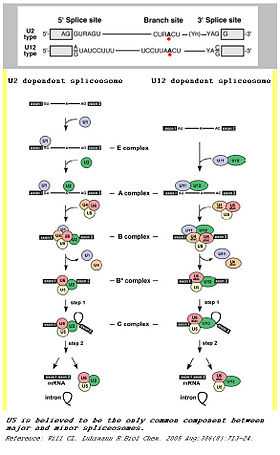
Spliceosomes are a major component of an integral step in eukaryotic precursor messenger RNA maturation. A mistake in even a single nucleotide can be devastating to the cell, and a reliable, repeatable method of RNA processing is necessary to ensure cell survival. The spliceosome is a large, protein-RNA complex that consists of five small nuclear RNAs (U1, U2, U4, U5, and U6) and over 150 proteins. The snRNAs, along with their associated proteins, form ribonucleoprotein complexes (snRNPs), which bind to specific sequences on the pre-mRNA substrate.[3] This intricate process results in two sequential transesterification reactions. These reactions will produce a free lariat intron and ligate two exons to form a mature mRNA. There are two separate classes of spliceosomes. The major class, which is far more abundant in eukaryotic cells, splices primarily U2-type introns. The initial step of splicing is the bonding of the U1 snRNP and its associated proteins to the 5’ splice end to the hnRNA. This creates the commitment complex which will constrain the hnRNA to the splicing pathway. [4] Then, U2 snRNP is recruited to the spliceosome binding site and forms complex A. [5] U2 snRNP changes the confirmation of the hnRNA-snRNP complex, exposing the nucleotide favorably for splicing. Following the confirmation change, the U4/U5/U6 tri-snRNP complex binds to complex A to form the structure known as complex B. After rearrangement, complex C is formed, and the spliceosome is active for catalysis. [6]
In addition to this main spliceosome complex, there exists a much less common (~1%) minor spliceosome. This complex comprises U11, U12, U4atac, U6atac and U5 snRNPs. These snRNPs are functional analogs of the snRNPs used in the major spliceosome. The minor spliceosome splices U-12 type introns. The two types of introns mainly differ in their splicing sites: U2-type introns have GT-AG 5’ and 3’ splice sites while U12-type introns have AT-AC at their 5’ and 3’ ends. The minor spliceosome carries out its function through a different pathway from the major spliceosome.
U1 snRNA

U1 snRNP is the initiator of spliceosomal activity in the cell by base pairing with the hnRNA. In the major spliceosome, experimental data has shown that the U1 snRNP is present in equal stoichiometry with U2, U4, U5, and U6 snRNP. However, U1 snRNP's abundance in human cells is far greater than that of the other snRNPs.[7] Through U1 snRNA gene knockdown in HeLa cells, studies have shown the U1 snRNA holds great importance for cellular function. When U1 snRNA genes were knocked out, genomic microarrays showed an increased accumulation of unspliced pre-mRNA. [8] In addition, the knockout was shown to cause premature cleavage and polyadenylation primarily in introns located near the beginning of the transcript. When other uridine based snRNAs were knocked out, this effect was not seen. Thus, U1 snRNA–pre-mRNA base pairing was shown to protect pre-mRNA from polyadenylation as well as premature cleavage. This special protection may explain the overabundance of U1 snRNA in the cell.
snRNPs and human disease
Through the study of small nuclear ribonucleoproteins (snRNPs) and small nucleolar (sno)RNPs we have been able to better understand many important diseases.
Spinal muscular atrophy - Mutations in the survival motor neuron-1 (SMN1) gene result in the degeneration of spinal motor neurons and severe muscle wasting. The SMN protein assembles Sm-class snRNPs, and probably also snoRNPs and other RNPs. [9] Spinal muscular atrophy affects up to 1 in 6,000 people, and is the second leading cause of neuromuscular disease, after Duchenne muscular dystrophy. [10]
Dyskeratosis congenital – Mutations in the assembled snRNPs are also found to be a cause of dyskeratosis congenital, a rare syndrome that presents by abnormal changes in the skin, nails and mucous membrane. Some ultimate effects of this disease include bone-marrow failure as well as cancer. This syndrome has been shown to arise from mutations in multiple genes, including dyskerin, telomerase RNA and telomerase reverse transcriptase. [11]
Prader–Willi syndrome - This syndrome affects as many as 1 in 12,000 people and has a presentation of extreme hunger, cognitive and behavioural problems, poor muscle tone and short stature.[12] The syndrome has been linked to the deletion of a region of paternal chromosome 15 that is not expressed on the maternal chromosome. This region includes a brain-specific snRNA that targets the serotonin-2C receptor mRNA.
References
- ↑ A. Gregory Matera, Rebecca M. Terns & Michael P. Terns. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNA. Nature Reviews Molecular Cell Biology 8, 209-220 (March 2007)
- ↑ Kiss, Tamás (2004-12-01). "Biogenesis of small nuclear RNPs". Journal of Cell Science 117 (25): 5949–5951. doi:10.1242/jcs.01487. ISSN 0021-9533. Retrieved 2013-07-24.
- ↑ Zhuojun Guo, Krishanthi S. Karunatilaka, and David Rueda. Single Molecule Analysis of Protein Free U2/U6 snRNAs. Nat Struct Mol Biol. 2009 November; 16(11): 1154–1159. Published online 2009 November 1.
- ↑ Legrain P, Seraphin B, Rosbash M (1 September 1988). "Early commitment of yeast pre-mRNA to the spliceosome pathway". Mol. Cell. Biol. 8 (9): 3755–60. PMC 365433
- ↑ Newby M. I. and Greenbaum, N. L. (2002). "Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine". Nature Structural Biology 9 (12): 958–65
- ↑ Burge, C.B., et al. (1999). "Splicing precursors to mRNAs by the spliceosomes". In Gesteland, R.F., Cech, T.R., Atkins, J.F.. The RNA World. Cold Spring Harbor Lab. Press. pp. 525–60.
- ↑ Baserga, S. J. & Steitz, J. A. in The RNA World (eds Gesteland, R. F. & Atkins, J. F.) 359–381 (Cold Spring Harbor Laboratory Press, 1993).
- ↑ Daisuke Kaida, Michael G. Berg, et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 468, 664–668 (2 December 2010).
- ↑ Matera, A. G. & Shpargel, K. B. Pumping RNA: nuclear bodybuilding along the RNP pipeline. Curr. Opin. Cell Biol. 18, 317–324 (2006)
- ↑ (Sarnat HB. Spinal muscular atrophies. In: Kliegman RM, Behrman RE, Jenson HB, Stanton BF. Nelson Textbook of Pediatrics. 19th ed. Philadelphia, Pa: Elsevier; 2011:chap 604.2.)
- ↑ (Wattendorf, D. J. & Muenke, M. Prader–Willi syndrome. Am. Fam. Physician 72, 827–830 (2005).)
- ↑ (Cooke DW, Divall SA, Radovick S. Normal and aberrant growth. In: Melmed S, ed. Williams Textbook of Endocrinology. 12th ed. Philadelphia, Pa: Saunders Elsevier; 2011:chap 24.)
See also
External links
- Small Nucleolar RNA at the US National Library of Medicine Medical Subject Headings (MeSH)
- Small Nuclear RNA at the US National Library of Medicine Medical Subject Headings (MeSH)
| ||||||||||||||||||||||||||||