Small hairpin RNA
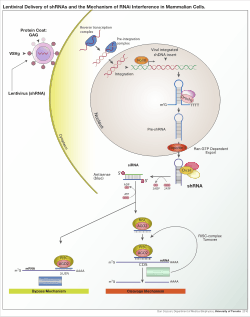
A small hairpin RNA or short hairpin RNA (shRNA) is a sequence of RNA that makes a tight hairpin turn that can be used to silence target gene expression via RNA interference (RNAi). Expression of shRNA in cells is typically accomplished by delivery of plasmids or through viral or bacterial vectors. The promoter choice is essential to achieve robust shRNA expression. At first, polymerase III promoters such as U6 and H1 were used; however, these promoters lack spatial and temporal control.[2] As such, there has been a shift to using polymerase II promoters to regulate expression of shRNA. shRNA is an advantageous mediator of RNAi in that it has a relatively low rate of degradation and turnover. However, shRNA is disadvantageous in that it requires use of an expression vector which can pose safety concerns.
Delivery
Expression of shRNA in cells can be obtained by delivery of plasmids or through viral or bacterial vectors.
Delivery of plasmids to cells through transfection to obtain shRNA expression can be accomplished using commercially available reagents in vitro. However, this method is not applicable in vivo and thus has limited utility.
Use of a bacterial vector to obtain shRNA expression in cells is a relatively recent approach. It builds off research showing that recombinant Escherichia coli, containing a plasmid with shRNA, fed to mice can knock-down target gene expression in the intestinal epithelium.[3] Interestingly, this approach was used in 2012 in clinical trials to try to treat patients with Familial Adenomatous Polyposis.
A variety of viral vectors can be used to obtain shRNA expression in cells including adeno-associated viruses (AAVs), adenoviruses, and lentiviruses. With adeno-associated viruses and adenoviruses, the genomes remain episomal. This is advantageous as insertional mutagenesis is avoided. It is disadvantageous in that the progeny of the cell will lose the virus quickly through cell division unless the cell divides very slowly. AAVs differ from adenoviruses in that the viral genes have been removed and they have diminished packing capacity. Lentiviruses integrate into sections of transcriptionally active chromatin and are thus passed on to progeny cells. With this approach there is increased risk of insertional mutagenesis; however, the risk can be reduced by using an integrase-deficient lentivirus.[4]
Mechanism of action
Once the vector has integrated into the host genome, the shRNA is then transcribed in the nucleus by polymerase II or polymerase III depending on the promoter choice. This product mimics pri-microRNA (pri-miRNA) and is processed by Drosha. The resulting pre-shRNA is exported from the nucleus by Exportin 5. This product is then processed by dicer and loaded into the RNA-induced silencing complex (RISC). The sense (passenger) strand is degraded. The antisense (guide) strand directs RISC to mRNA that has a complementary sequence. In the case of perfect complementarity, RISC cleaves the mRNA. In the case of imperfect complementarity, RISC represses translation of the mRNA. In both of these cases, the shRNA leads to target gene silencing.
Applications in gene therapy
Due to the ability of shRNA to provide specific, long-lasting, gene silencing there has been great interest in using shRNA for gene therapy applications. Three examples of shRNA-based therapies are discussed below.
Gradalis, Inc. developed the FANG vaccine, which is used in treatment of advanced cancers. FANG relies on a bifunctional shRNA (bi-shRNA) against the immunosuppressive transforming growth factors (TGF) β1 and β2.[5] Autologous tumor cells were harvested from patients and a plasmid encoding the bifunctional shRNA and granulocyte-macrophage colony-stimulating factor (GMCSF) was introduced ex vivo through electroporation. These cells were later irradiated and injected back into the patient.
Marina Biotech developed CEQ508 which is used to treat Familial Adenomatous Polyposis. CEQ508 uses a bacterial vector to deliver shRNA against β-catenin.
Gradalis, Inc. developed bifunctional shRNA-STMN1 (pbi-shRNA STMN1), which is used to treat advanced and/or metastatic cancers. This pbi-shRNA STMN1 is against stathmin 1 and is delivered intratumorally through bilamellar invaginated vesicle (BIV) lipoplex (LP) technology.
Several challenges typically confront shRNA-based therapeutics. The most significant challenge is delivery. shRNA is typically delivered through use of a vector, and although they are generally efficient, they pose significant safety concerns. In particular, viral based gene therapy approaches have proved dangerous in past clinical trials. Potential oversaturation of RISC is also a problem. If the shRNA is expressed at levels that are too high the cell might not be able to correctly process the endogenous RNA which could cause significant problems. Another challenge is the possibility that the patient might mount an immune response against the therapy.[6] Finally, there might be off-target effects and the shRNA could silence other unintended genes. In developing successful new shRNA-based therapeutics, all of these challenges must be taken into account.
References
- ↑ Macrae I, Zhou K, Li F, Repic A, Brooks A, Cande W, Adams P, Doudna J (2006). "Structural basis for double-stranded RNA processing by dicer". Science 311 (5758): 195–8. doi:10.1126/science.1121638. PMID 16410517.
- ↑ Wang, Zhaohui; Rao, Donald D.; Senzer, Neil; Nemunaitis, John (2011). "RNA Interference and Cancer Therapy". Pharmaceutical Research 28 (12): 2983–2995. doi:10.1007/s11095-011-0604-5. ISSN 0724-8741.
- ↑ Xiang, Shuanglin; Fruehauf, Johannes; Li, Chiang J (2006). "Short hairpin RNA–expressing bacteria elicit RNA interference in mammals". Nature Biotechnology 24 (6): 697–702. doi:10.1038/nbt1211. ISSN 1087-0156.
- ↑ Lombardo, Angelo; Genovese, Pietro; Beausejour, Christian M; Colleoni, Silvia; Lee, Ya-Li; Kim, Kenneth A; Ando, Dale; Urnov, Fyodor D; Galli, Cesare; Gregory, Philip D; Holmes, Michael C; Naldini, Luigi (2007). "Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery". Nature Biotechnology 25 (11): 1298–1306. doi:10.1038/nbt1353. ISSN 1087-0156.
- ↑ Senzer, Neil; Barve, Minal; Kuhn, Joseph; Melnyk, Anton; Beitsch, Peter; Lazar, Martin; Lifshitz, Samuel; Magee, Mitchell; Oh, Jonathan; Mill, Susan W; Bedell, Cynthia; Higgs, Candice; Kumar, Padmasini; Yu, Yang; Norvell, Fabienne; Phalon, Connor; Taquet, Nicolas; Rao, Donald D; Wang, Zhaohui; Jay, Chris M; Pappen, Beena O; Wallraven, Gladice; Brunicardi, F Charles; Shanahan, David M; Maples, Phillip B; Nemunaitis, John (2011). "Phase I Trial of “bi-shRNAifurin/GMCSF DNA/Autologous Tumor Cell” Vaccine (FANG) in Advanced Cancer". Molecular Therapy 20 (3): 679–686. doi:10.1038/mt.2011.269. ISSN 1525-0016. PMC 3293620. PMID 22186789.
- ↑ Silencing or stimulation? siRNA delivery and the immune system 2. 2011. pp. 77–96. doi:10.1146/annurev-chembioeng-061010-114133. PMID 22432611.
| ||||||||||||||||||||||||||||
