Second-harmonic imaging microscopy
Second-harmonic imaging microscopy (SHIM) is based on a nonlinear optical effect known as second-harmonic generation (SHG). SHIM has been established as a viable microscope imaging contrast mechanism for visualization of cell and tissue structure and function. A second-harmonic microscope obtains contrasts from variations in a specimen’s ability to generate second-harmonic light from the incident light while a conventional optical microscope obtains its contrast by detecting variations in optical density, path length, or refractive index of the specimen. SHG requires intense laser light passing through a material with a noncentrosymmetric molecular structure. Second-harmonic light emerging from an SHG material is exactly half the wavelength (frequency doubled) of the light entering the material. While two-photon-excited fluorescence (TPEF) is also a two photon process, TPEF loses some energy during the relaxation of the excited state, and SHG is energy conserving. Typically, an inorganic crystal is used to produce SHG light such as lithium niobate (LiNbO3), potassium titanyl phosphate (KTP = KTiOPO4), and lithium triborate (LBO = LiB3O5). Though SHG requires a material to have specific molecular orientation in order for the incident light to be frequency doubled, some biological materials can be highly polarizable, and assemble into fairly ordered, large noncentrosymmetric structures. Biological materials such as collagen, microtubules, and muscle myosin can produce SHG signals. The SHG pattern is mainly determined by the phase matching condition. A common setup for an SHG imaging system will have a laser scanning microscope with a titanium sapphire mode-locked laser as the excitation source. The SHG signal is propagated in the forward direction. However, some experiments have shown that objects on the order of about a tenth of the wavelength of the SHG produced signal will produce nearly equal forward and backward signals.
Advantages
SHIM offers several advantages for live cell and tissue imaging. SHG does not involve the excitation of molecules like other techniques such as fluorescence microscopy therefore, the molecules shouldn’t suffer the effects of phototoxicity or photobleaching. Also, since many biological structures produce strong SHG signals, the labeling of molecules with exogenous probes is not required which can also alter the way a biological system functions. By using near infrared wavelengths for the incident light, SHIM has the ability to construct three dimensional images of specimens by imaging deeper into thick tissues.
History
Before SHG was used for imaging, the first demonstration of SHG was performed in 1961 by P. A. Franken, G. Weinriech, C. W. Peters, and A. E. Hill at the University of Michigan, Ann Arbor using a quartz sample.[1] In 1968, SHG from interfaces was discovered by Bloembergen and has since been used as a tool for characterizing surfaces and probing interface dynamics. In 1971, Fine and Hansen reported the first observation of SHG from biological tissue samples. In 1974, Hellwarth and Christensen first reported the integration of SHG and microscopy by imaging SHG signals from polycrystalline ZnSe. In 1977, Colin Sheppard imaged various SHG crystals with a scanning optical microscope. The first biological imaging experiments were done by Freund in 1986 to study the orientation of collagen fibers in rat tail tendon. In 1993, Lewis examined the second-harmonic response of styryl dyes in electric fields. He also showed work on imaging live cells.
In 2010 SHG was extended to whole-animal in vivo imaging.[2][3]
Applications
SHG polarization anisotropy can be used to determine the orientation and degree of organization of proteins in tissues since SHG signals have well-defined polarizations. By using the anisotropy equation:
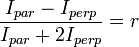
and acquiring the intensities of the polarizations in the parallel and perpendicular directions. A high  value indicates an anisotropic orientation whereas a low
value indicates an anisotropic orientation whereas a low  value indicates an isotropic structure. In work done by Campagnola and Loew, it was found that collagen fibers formed well-aligned structures with an
value indicates an isotropic structure. In work done by Campagnola and Loew, it was found that collagen fibers formed well-aligned structures with an  value.
value.
It has also been used to prove that backpropagating action potentials invade dendritic spines without voltage attenuation, establishing a sound basis for future work on Long-term potentiation. Its use here was that it provided a way to accurately measure the voltage in the tiny dendritic spines with an accuracy unattainable with standard two-photon microscopy.[4]
See also
Sources
- Campagnola, Paul J.; Clark, Heather A.; Mohler, William A.; Lewis, Aaron; Loew, Leslie M. (2001). "Second harmonic imaging microscopy of living cells". Journal of Biomedical Optics (SPIE) 6 (3): 277–286. Bibcode:2001JBO.....6..277C. doi:10.1117/1.1383294.
- Campagnola, Paul J.; Loew, Leslie M (2003). "Second-harmonic imaging microscopy for visualizing biomolecular arrays in cells, tissues and organisms". Nature Biotechnology 21: 1356–1360. doi:10.1038/nbt894.
- P. Stoller, K.M. Reiser, P.M. Celliers, & A.M. Rubenchik, “Polarization-modulated second harmonic generation in collagen.” Biophys. J. 82, 3330–3342 (2002)
- M. Han, G. Giese, and J. F. Bille, “Second harmonic generation imaging of collagen fibrils in cornea and sclera,” Opt. Express 13, 5791–5797 (2005)
- ↑ Franken, Peter; Weinreich, G; Peters, CW; Hill, AE (1961). "Generation of Optical Harmonics". Physical Review Letters 7 (4): 118–119. Bibcode:1961PhRvL...7..118F. doi:10.1103/PhysRevLett.7.118.
- ↑ Cohen, B. E. (2010). "Biological imaging: Beyond fluorescence". Nature 467 (7314): 407. Bibcode:2010Natur.467..407C. doi:10.1038/467407a. PMID 20864989.
- ↑ Pantazis, P.; Maloney, J.; Wu, D.; Fraser, S. (2010). "Second harmonic generating (SHG) nanoprobes for in vivo imaging". Proceedings of the National Academy of Sciences of the United States of America 107 (33): 14535–14540. Bibcode:2010PNAS..10714535P. doi:10.1073/pnas.1004748107. PMC 2930484. PMID 20668245.
- ↑ Nuriya, Mutsuo; Jiang, Jiang; Nemet, Boaz; Eisenthal, Kenneth B.; Rafael, Yuste (2006). "Imaging membrane potential in dendritic spines". PNAS 103 (3): 786–790. Bibcode:2006PNAS..103..786N. doi:10.1073/pnas.0510092103.
| |||||||||||||||||
