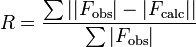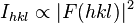R-factor (crystallography)
In crystallography, the R-factor (sometimes called residual factor or reliability factor or the R-value or RWork) is a measure of the agreement between the crystallographic model and the experimental X-ray diffraction data. In other words, it is a measure of how well the refined structure predicts the observed data.[1] The value is also sometimes called the discrepancy index, as it mathematically describes the difference between the experimental observations and the ideal calcluated values.[2] It is defined by the following equation:
where F is the so-called structure factor and the sum extends over all the reflections measured and their calculated counterparts respectively. The structure factor is closely related to the intensity of the reflection it describes:
The absolute range of values is zero to one, with a large R-factor indicating a poorly refined, low-resolution model or a disordered crystal. For large molecules, R-factor usually ranges between 0.6 (when comparing a random set of reflections with a given model) and 0.2 (for example for a well refined macro-molecular model at a resolution of 2.5 Ångström). Small molecules (up to 300 atoms) usually form more ordered crystals than large molecules, it is possible to attain lower R-factors. In the Cambridge Structural Database more than 95% of the 500,000+ crystals have an R-factor lower than 0.15 and 9.5% have an R-factor lower than 0.03.
Crystallographers also use the Free R-Factor (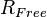 ) [3] to describe the quality of a model.
) [3] to describe the quality of a model.
The quantities 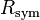 and
and 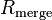 are similarly used to describe the internal agreement of measurements in a crystallographic data set.
are similarly used to describe the internal agreement of measurements in a crystallographic data set.
References
- ↑ Morris AL, MacArthur MW, Hutchinson EG, Thornton JM (April 1992). "Stereochemical quality of protein structure coordinates". Proteins 12 (4): 345–64. doi:10.1002/prot.340120407. PMID 1579569.
- ↑ "R factor". International Union of Crystallography. Retrieved 2013-12-13.
- ↑ Brunger AT (January 1992). "Free R value: a novel statistical quantity for assessing the accuracy of crystal structures". Nature 355 (6359): 472–475. Bibcode:1992Natur.355..472B. doi:10.1038/355472a0. PMID 18481394.