Pyrimidine dimer

Pyrimidine dimers are molecular lesions formed from thymine or cytosine bases in DNA via photochemical reactions.[1][2] Ultraviolet light induces the formation of covalent linkages by reactions localized on the C=C double bonds.[3] In dsRNA, uracil dimers may also accumulate as a result of UV radiation. Two common UV products are cyclobutane pyrimidine dimers (CPDs, including thymine dimers) and 6,4 photoproducts. These premutagenic lesions alter the structure of DNA and consequently inhibit polymerases and arrest replication. Dimers may be repaired by photoreactivation or nucleotide excision repair, but unrepaired dimers are mutagenic. Pyrimidine dimers are the primary cause of melanomas in human beings. [citation needed]
Types of dimers
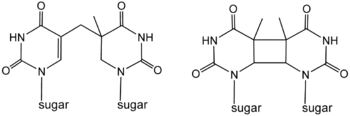
6,4-photoproducts, or 6,4 pyrimidine-pyrimidones, occur at one third the frequency of CPDs but are more mutagenic.[7] Spore photoproduct lyase provides another enzymatic pathway for repair of thymine photodimers.[8]
Mutagenesis
Translesion polymerases frequently introduce mutations at pyrimidine dimers, both in prokaryotes (SOS mutagenesis) and in eukaryotes. Although the thymine-thymine CPDs (thymine dimers) are the most frequent lesions caused by UV light, translesion polymerases are biased toward introduction of As, so that TT dimers are often replicated correctly. On the other hand, any C involved in CPDs is prone to be deaminated, inducing a C to T transition.[9]
DNA repair

Pyrimidine dimers introduce local conformational changes in the DNA structure, which allow recognition of the lesion by repair enzymes.[10] In most organisms (excluding placental mammals such as humans) they can be repaired by photoreactivation.[11] Photoreactivation is a repair process in which photolyase enzymes directly reverse CPDs via photochemical reactions. Lesions on the DNA strand are recognized by these enzymes, followed by the absorption of light wavelengths >300 nm (i.e. fluorescent and sunlight). This absorption enables the photochemical reactions to occur, which results in the elimination of the pyrimidine dimer, returning it to its original state.[12]
Nucleotide excision repair is a more general mechanism for repair of lesions. This process excises the CPD and synthesizes new DNA to replace the surrounding region in the molecule.[12] Xeroderma pigmentosum is a genetic disease in humans in which the nucleotide excision repair process is lacking, resulting in skin discolouration and multiple tumours on exposure to UV light. Unrepaired pyrimidine dimers in humans may lead to melanoma.[13]
References
- ↑ David S. Goodsell (2001). "The Molecular Perspective: Ultraviolet Light and Pyrimidine Dimers". The Oncologist 6 (3): 298–299. doi:10.1634/theoncologist.6-3-298. PMID 11423677.
- ↑ E. C. Friedberg, G. C. Walker, W. Siede, R. D. Wood, R. A. Schultz and T. Ellenberger (2006). DNA repair and mutagenesis. Washington: ASM Press. p. 1118. ISBN 978-1-55581-319-2.
- ↑ S. E. Whitmore, C. S. Potten, C. A. Chadwick, P. T. Strickland, W. L. Morison (2001). "Effect of photoreactivating light on UV radiation-induced alterations in human skin". Photodermatol. Photoimmunol. Photomed. 17 (5): 213–217. doi:10.1034/j.1600-0781.2001.170502.x. PMID 11555330.
- ↑ R. B. Setlow (1966). "Cyclobutane-Type Pyrimidine Dimers in Polynucleotides". Science 153 (3734): 379–386. doi:10.1126/science.153.3734.379. PMID 5328566.
- ↑ Expert reviews in molecular medicine (2 December 2002). "Structure of the major UV-induced photoproducts in DNA.". Cambridge University Press.
- ↑ Christopher Mathews and K.E. Van Holde (1990). Biochemistry (2nd ed.). Benjamin Cummings Publication. p. 1168. ISBN 978-0-8053-5015-9.
- ↑ Van Holde, K. E.; Mathews, Christopher K. (1990). Biochemistry. Menlo Park, Calif: Benjamin/Cummings Pub. Co. ISBN 0-8053-5015-2.
- ↑ Jeffrey M. Buis, Jennifer Cheek, Efthalia Kalliri, and Joan B. Broderick (2006). "Characterization of an Active Spore Photoproduct Lyase, a DNA Repair Enzyme in the Radical S-Adenosylmethionine Superfamily". Journal of Biological Chemistry 281 (36): 25994–26003. doi:10.1074/jbc.M603931200. PMID 16829680.
- ↑ J. H. Choi, A. Besaratinia ,D. H. Lee, C. S. Lee, G. P. Pfeifer (2006). "The role of DNA polymerase iota in UV mutational spectra". Mutat. Res. 599 (1–2): 58–65. doi:10.1016/j.mrfmmm.2006.01.003. PMID 16472831.
- ↑ Kemmink Johan, Boelens Rolf, Koning Thea M.G., Kaptein Robert, Van , der Morel Gijs A., Van Boom Jacques H. (1987). "Conformational Changes in the oligonucleotide duplex d(GCGTTGCG)*d(GCGAAGCG) induced by formation of a cis-syn thymine dimer". European Journal of Biochemistry 162: 31–43. doi:10.1111/j.1432-1033.1987.tb10538.x. PMID 3028790.
- ↑ Essen LO, Klar T (2006). "Light-driven DNA repair by photolyases". Cell Mol Life Sci 63 (11): 1266–77. doi:10.1007/s00018-005-5447-y. PMID 16699813.
- ↑ 12.0 12.1 Friedberg, Errol C. (23 January 2003) "DNA Damage and Repair". Nature 421, 436-439. doi:10.1038/nature01408
- ↑ Vink Arie A., Roza Len (2001). "Biological consequences of cyclobutane pyrimidine dimers". Journal of Photochemistry and Photobiology B: Biology 65 (2–3): 101–104. doi:10.1016/S1011-1344(01)00245-7.