Proteus penneri
| Proteus penneri | |
|---|---|
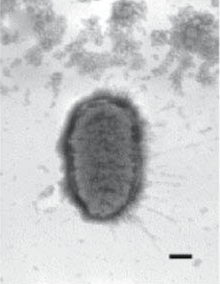 | |
| Electron micrograph of Proteus penneri. Bar represents 200nm. | |
| Scientific classification | |
| Kingdom: | Bacteria |
| Phylum: | Proteobacteria |
| Class: | Gamma Proteobacteria |
| Order: | Enterobacteriales |
| Family: | Enterobacteriaceae |
| Genus: | Proteus |
| Species: | P. penneri |
| Binomial name | |
| Proteus penneri Hickman et al. 1982 | |
Proteus penneri is a Gram-negative, facultatively anaerobic, rod shaped bacterium.[1] It is an invasive pathogen[2] and a cause of nosocomial infections of the urinary tract or in open wounds.[3] Pathogens have been isolated mainly from the urine of patients with abnormalities in the urinary tract, and from stool.<ref name="pmid11023955" [">O'Hara CM, Brenner FW, Miller JM (2000). "Classification, identification, and clinical significance of Proteus, Providencia, and Morganella.". Clin Microbiol Rev 13 (4): 534–46. PMC 88947. PMID 11023955.</ref> P. penneri strains are naturally resistant to numerous antibiotics, including penicillin G, amoxicillin, cephalosporins, oxacillin, and most macrolides, but are naturally sensitive to aminoglycosides, carbapenems, aztreonam, quinolones, sulphamethoxazole and co-trimoxazole. Isolates of P. penneri have been found to be multi-drug resistant (MDR) with resistance to 6-8 drugs. β-lactamase production has also been identified in some isolates.[4]
History
The Proteus penneri group of bacteria was named in 1982. It reclassified a group of strains formerly known as Proteus vulgaris biogroup 1.[5] In 1978, Brenner et al. showed through DNA hybridization studies that P.vulgaris was a heterogenous species.[6] In 1981, Hickman et al conducted experiments on 20 indole-negative strains previously grouped with P.vulgaris and demonstrated the existence of three P. vulgaris biogroups. Proteus vulgaris biogroup 1, or indole-negative P. vulgaris, was distinguished as a new species within the Proteus genus in 1982.[1] The new species was named Proteus Penneri in honor of John Penner, a Canadian microbiologist who contributed greatly to the study of the three genera of Proteeae.[7]
| Date | Author | Event |
|---|---|---|
| 1885 | Hauser | Described the genus Proteus and species P. mirabilis, P. vulgaris, and P. zenkeri. |
| 1919 | Wenner and Rettger | Separated P. vulgaris and P. mirabilis on the basis of sugar fermentation. |
| 1978 | Brenner et al. | Defined P. vulgaris biogroup 1. |
| 1982 | Hickman et al. | Distinguished P. vulgaris biogroup 1 as a new species named Proteus penneri. |
Lab identification and differentiation
Extended biochemical tests have characterized P.penneri as being uniformly salicin negative. The inability to produce ornithine decarboxylase differentiates P.penneri from another indole-negative Proteus species P.mirabilis.[2] P. penneri isolates are non-fermenters of salicin and non-utilizers of citrate but acidify sucrose and maltose.[4] Other chief characteristics of this species that enable its differentiation from other Proteus species include failure to acidify esculin, failure to produce hydrogen sulfide on triple sugar iron agar, and resistance to chloramphenicol.[8] The resistance of P. penneri to cefuroxime and the marked inhibitory activity of cefoxitin against this species also distinguishes P. penneri from the other Proteeae.[9] Similar to other Proteus species, P.penneri has a cell-bound hemolytic factor, which has been shown to facilitate penetration of the organism into cultured Vero cells without any cytotoxic effects. It also has a filterable cytotoxic alpha-hemolysin rarely found in other Proteus species.[6] A highly active urease produced by P.penneri may also have a role in the establishment of an infectious process.[8]
The application of molecular techniques such as the polymerase chain reaction (PCR) to produce DNA fingerprints and other 16S ribosomal RNA gene (ribotyping) methods of strain analysis have been employed to differentiate P.penneri from P.vulgaris and P.mirabilis strains. The RAPD technique, fundamentally a DNA fingerprinting method, has exposed a substantial DNA diversity among P.Penneri strains, a characteristic that remained unidentified by other methods.[10]
| Test | Result |
|---|---|
| Microscopic morphology | Gram-negative rods |
| Hemolysis (sheep blood agar) | Beta |
| Urease | Positive |
| Indole production | Negative |
| Esculin hydrolysis | Negative |
| Acid from Maltose | Positive |
| Acid from Sucrose | Positive |
| Citrate utilization | Negative |
| Ornithine decarboxylase | Negative |
| Hydrogen sulfide production | Negative |
Subtypes
The lipopolysaccharide (LPS) core region of Proteus penneri strains contains higher structural variability than that observed in other representatives of Enterobacteriaceae.[11] These differences have been utilized to cluster P. penneri strains into serogroups based on their agglutinating activity when mixed with antibodies directed against specific species of LPS molecules.[12] Presently, 15 O-serogroups have been proposed for P. penneri based on the chemical structure of the O-specific polysaccharide chain (O-antigen) of the lipopolysaccharide.[13][14][15] Certain LPS epitopes have been examined to determine their function in antigenic specificity. The particular groups on the oligosaccharides found to play a dominant role in the specificity of P.penneri LPS are the amide of D-galacturonic acid with L-lysine α-D-GalA-(L-Lys) (and the amide of D-galacturonic acid with L-threonine α-D-GalA- [L-Thr]), respectively.[12]
Isolation
The occurrence of P.penneri organisms in the normal intestine accounts for their higher frequency in in urinary tract infections and for their role as opportunistic invaders after surgery.[16] P. penneri is absent from the intestines of livestock.[10] Optimum growth conditions for P.penneri is achieved at 37°C, which mirrors the intestinal niche colonized by many of these bacteria. Certain strains of P. penneri can differentiate into elongated hyper-flagelled cells during development on solid media, resulting in the surface translocation event identified as “swarming”. Swarming motility makes it difficult to isolate single colonies for further study.[17]
Incidence and Epidemiology
It is unknown what proportion of P.penneri strains have been isolated in clinical specimens.[9] Since P.penneri was only recently recognized as a new species, many bacteriologists are either generally unaware of P. penneri, or have made limited attempts to discover its clinical significance. Thus, reports on the isolation of P. penneri from infected patients is limited.[2] Although increasing numbers of laboratories are now identifying P.penneri strains, the number reported in susceptibility studies are relatively small for a general assessment of the incidence of the species.[9] Likewise, the epidemiology of P.penneri is also unknown for the aforementioned reasons.
Clinical significance
Documented human clinical infections caused by Proteus penneri have been limited to the urinary tract and to wounds of the abdomen, groin, neck, and ankle.[8]Proteus penneri is isolated from individuals in long-term care facilities, hospitals and from patients who are immunocompromised or suffering from underlying disease. In a study by Müller, P. penneri was isolated significantly more often from stools of patients with diarrheal disease than from healthy patients, leading the author to speculate that P. penneri may play a role in some diarrheal disease.[18] The invasive potential of this microorganism has also been demonstrated in a case of P. penneri bacteremia and concomitant subcutaneous thigh abscess in a neutropenic patient with acute lymphocytic leukemia [8] and in nosocomial urosepsis in a diabetic patient from whom the organism was also subsequently isolated from bronchoalveolar lavage fluid and a pulmonary catheter tip.[19] Furthermore, in an experiment conducted in India, P.penneri strains were isolated as the sole pathogen in all patients having underlying disease; post-operatively. Most isolates of P.penneri from the experiment were found to be multi-drug resistant including resistance to amoxy-clavulanic acid combination.[2] In another study conducted in Germany, P. penneri was found to be more resistant to the penicillins and cephalosporins than P. mirabilis and mostly in patients with urogenital infections.[4] Moreover, the urease enzyme of P. penneri is thought to be a significant cause of kidney stone formation. Consistent with this belief, the organism has been isolated from the center of a stone removed from a patient with persistent P. penneri bacteriuria.[20] This data substantiates the need for species level identification of P.penneri in the clinical setting.
Virulence Factors
Several virulence factors of P.penneri can make infections from this invasive pathogen more pronounced, persistent, and harder to eradicate.[2] These include adherence due to the presence of fimbriae or afimbrial adhesins, invasiveness, swarming phenomenon, hemolytic activity, urea hydrolysis, proteolysis, and endotoxicity.[21] Swarming motility is the coordinated translocation of a bacterial population driven by flagellar rotation in film or on fluid surfaces.[22] 6. An emerging concept in microbiology, the fundamental role of swarming motility remains unknown. However, it has been observed to be correlated with an elevated resistance to certain antibiotics.[23] Production of IgA proteolytic enzymes has also been reported in Proteus penneri.[24] Secretory immunoglobulins of the IgA class are produced by mucous tissue and are particularly resistant to enzymatic breakdown by proteases. The ability to degrade a host’s secretory IgA may provide P.penneri with an advantage by permitting it to evade the host immune response, therefore gaining valuable time for the bacterium to establish a foothold for infection.However, the major mechanism of antimicrobial resistance is caused by hyperproduction of the chromosomally encoded β-lactamase, sometimes via plasmids. These inducible β-lactamases will hydrolyze primary and extended-spectrum penicillins and cephalosporins,[25] thus making P. penneri strains naturally resistant to penicillin G, amoxicillin, cephalosporins (i.e. cefaclor, cefazoline, cefuroxime and cefdinir), oxacillin, and most macrolides.[2]
Susceptibility Profile
Most isolated Proteus penneri strains are multi-drug resistant (MDR), with 12 being the highest drug resistance number reported.[2] P.penneri has a distinguishing susceptibility profile, based on the production of the chromosomally-induced Beta-lactamase HugA. HugA determines resistance to aminopenicillins and first generation and second generation cephalosporins,including cefuroxime. However HugA does not affect cephamycins or carbapenems and is inhibited by clavulanic acid. Similar to other Proteus species, P.penneri is resistant to tetracyclines and nitrofurantoin.[26]
Presently, all tested strains of Proteus penneri have been found to be highly susceptible to:[27]
- ceftizoxime
- ceftazidime
- moxalactam
- cefoxitin
- gentamicin
- tobramycin
- netilmicin
Most strains with a few exceptions are also susceptible to:
All tested Proteus penneri strains have been found resistant to:
- penicillin
- ampicillin
- tetracycline
- chloramphenicol
- co-trimoxazole
Treatment
There is limited information on the treatment of P.penneri infections. However, the use of the following aminoglycosides: gentamicin, tobramycin, netilmicin, and amikacin have been indicated as possible drugs of choice for the treatment of systemic infections caused by susceptible P. penneri strains. In vitro studies of ceftizoxime, ceftazidime, moxalactam, and cefoxitin suggest that these agents also may prove to one day be clinically useful in treating infections caused by P. penneri.[27]
See also
References
- ↑ 1.0 1.1 Hickman, FW; Steigerwalt, AG, Farmer JJ, 3rd, Brenner, DJ (June 1982). "Identification of Proteus penneri sp. nov., formerly known as Proteus vulgaris indole negative or as Proteus vulgaris biogroup 1.". Journal of clinical microbiology 15 (6): 1097–102. PMC 272260. PMID 7050147.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 Kishore J (2012). "Isolation, identification & characterization of Proteus penneri--a missed rare pathogen.". Indian J Med Res 135: 341–5. PMC 3361870. PMID 22561620.
- ↑ Kaistha, N; Bansal, N, Chander, J (July 2011). "Proteus penneri lurking in the intensive care unit: An important often ignored nosocomial pathogen.". Indian journal of anaesthesia 55 (4): 411–3. doi:10.4103/0019-5049.84842. PMC 3190523. PMID 22013265.
- ↑ 4.0 4.1 4.2 Stock I (2003). "Natural antibiotic susceptibility of Proteus spp., with special reference to P. mirabilis and P. penneri strains.". J Chemother 15 (1): 12–26. PMID 12678409.
- ↑ Krajden S, Fuksa M, Petrea C, Crisp LJ, Penner JL (1987). "Expanded clinical spectrum of infections caused by Proteus penneri.". J Clin Microbiol 25 (3): 578–9. PMC 266001. PMID 3571463.
- ↑ 6.0 6.1 Rozalski A, Kotełko K (1987). "Hemolytic activity and invasiveness in strains of Proteus penneri.". J Clin Microbiol 25 (6): 1094–6. PMC 269143. PMID 3597752.
- ↑
- ↑ 8.0 8.1 8.2 8.3 Engler HD, Troy K, Bottone EJ (1990). "Bacteremia and subcutaneous abscess caused by Proteus penneri in a neutropenic host.". J Clin Microbiol 28 (7): 1645–6. PMC 268005. PMID 2380386.
- ↑ 9.0 9.1 9.2 Piccolomini R, Cellini L, Allocati N, Di Girolamo A, Ravagnan G (1987). "Comparative in vitro activities of 13 antimicrobial agents against Morganella-Proteus-Providencia group bacteria from urinary tract infections.". Antimicrob Agents Chemother 31 (10): 1644–7. PMC 175007. PMID 3435110.
- ↑ 10.0 10.1 Costas M, Holmes B, Frith KA, Riddle C, Hawkey PM (1993). "Identification and typing of Proteus penneri and Proteus vulgaris biogroups 2 and 3, from clinical sources, by computerized analysis of electrophoretic protein patterns.". J Appl Bacteriol 75 (5): 489–98. PMID 8300450.
- ↑ Palusiak A, Sidorczyk Z (2010). "Characterization of epitope specificity of Proteus penneri 7 lipopolysaccharide core region.". Acta Biochim Pol 57 (4): 529–32. PMID 21060898.
- ↑ 12.0 12.1 Manos, J.; Belas, R. (2006). "The Genera Proteus, Providencia, and Morganella". The Prokaryotes. p. 245. doi:10.1007/0-387-30746-x_12. ISBN 978-0-387-25496-8.
- ↑ Cedzyński M, Knirel YA, Rózalski A, Shashkov AS, Vinogradov EV, Kaca W (1995). "The structure and serological specificity of Proteus mirabilis O43 O antigen.". Eur J Biochem 232 (2): 558–62. PMID 7556207.
- ↑ Sidorczyk Z., Zych K., Kołodziejska K., Drzewiecka D. and Zabłotni A. (2002) Progress in serological classification of further strains from genus Proteusand determination of epitopes and new serogroups. Second German-Polish-Russian Meeting on Bacterial Carbohydrates, Moscow, September 10–12, 2002.
- ↑ Zych K, Kowalczyk M, Knirel YA, Sidorczyk Z (2000). "New serogroups of the genus Proteus consisting of Proteus penneri strains only. Determination of some LPS epitopes responsible for specificity.". Adv Exp Med Biol 485: 339–44. doi:10.1007/0-306-46840-9_47. PMID 11109127.
- ↑ Krajden S, Fuksa M, Lizewski W, Barton L, Lee A (1984). "Proteus penneri and urinary calculi formation.". J Clin Microbiol 19 (4): 541–2. PMC 271113. PMID 6715521.
- ↑ Belas R, Schneider R, Melch M (1998). "Characterization of Proteus mirabilis precocious swarming mutants: identification of rsbA, encoding a regulator of swarming behavior.". J Bacteriol 180 (23): 6126–39. PMC 107696. PMID 9829920.
- ↑ Müller HE (1986). "Occurrence and pathogenic role of Morganella-Proteus-Providencia group bacteria in human feces.". J Clin Microbiol 23 (2): 404–5. PMC 268658. PMID 3517057.
- ↑ Latuszynski DK, Schoch P, Qadir MT, Cunha BA (1998). "Proteus penneri urosepsis in a patient with diabetes mellitus.". Heart Lung 27 (2): 146–8. PMID 9548071.
- ↑ Griffith DP, Musher DM, Itin C (1976). "Urease. The primary cause of infection-induced urinary stones.". Invest Urol 13 (5): 346–50. PMID 815197.
- ↑ Rózalski A, Kwil I, Torzewska A, Baranowska M, Staczek P (2007). "[Proteus bacilli: features and virulence factors].". Postepy Hig Med Dosw (Online) 61: 204–19. PMID 17507868.
- ↑ Fraser GM, Hughes C (1999). "Swarming motility.". Curr Opin Microbiol 2 (6): 630–5. PMID 10607626.
- ↑ Kim W, Killam T, Sood V, Surette MG (2003). "Swarm-cell differentiation in Salmonella enterica serovar typhimurium results in elevated resistance to multiple antibiotics.". J Bacteriol 185 (10): 3111–7. doi:10.1128/JB.185.10.3111-3117.2003. PMC 154059. PMID 12730171.
- ↑ Loomes LM, Senior BW, Kerr MA (1990). "A proteolytic enzyme secreted by Proteus mirabilis degrades immunoglobulins of the immunoglobulin A1 (IgA1), IgA2, and IgG isotypes.". Infect Immun 58 (6): 1979–85. PMC 258753. PMID 2111288.
- ↑ Swenson, JM; Hindler J A, Peterson L R. (1999). Murray, P.R., ed. Manual of clinical microbiology. Washington, D.C. pp. 1563–1577.
- ↑ Cantón R, Sánchez-Moreno MP, Morosini Reilly MI (2006). "[Proteus penneri].". Enferm Infecc Microbiol Clin. 24 Suppl 1: 8–13. PMID 17125662.
- ↑ 27.0 27.1 Fuksa M, Krajden S, Lee A (1984). "Susceptibilities of 45 clinical isolates of Proteus penneri.". Antimicrob Agents Chemother 26 (3): 419–20. PMC 176184. PMID 6508270.