Protein-fragment complementation assay
Protein-fragment complementation assay, or PCA, is a method for the identification of protein–protein interactions in biological systems. It is used in the field of proteomics. In the PCA, the proteins of interest ("Bait" and "Prey") are each covalently linked to incomplete fragments of a third protein (e.g. DHFR, which acts as a "reporter"). Interaction between the "bait" and the "prey" proteins brings the fragments of the "reporter" protein in close enough proximity to allow them to form a functional reporter protein whose activity can be measured. This principle can be applied to many different "reporter" proteins and is also the basis for the yeast two-hybrid system, an archetypical PCA assay.
Split protein assays
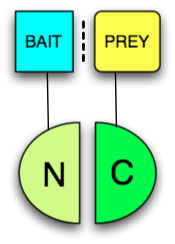
Any protein that can be split into two parts and reconstituted non-covalently may be used in a PCA. The two parts just have to be brought together by other interacting proteins fused to them (typically called "bait" and "prey" (see figure). The protein that produces a detectable readout is called "reporter". Usually enzymes which confer resistance to antibiotics, such as Dihydrofolate reductase or Beta-lactamase, or proteins that give colorimetric or fluorescent signals are used as reporters. When fluorescent proteins are reconstituted the PCA is called Bimolecular fluorescence complementation assay. The most popular PCAs utilize split versions of the following proteins:
- Dihydrofolate reductase (DHFR)[1]
- Beta-lactamase[2][3]
- Yeast Gal4 (as in the classical yeast two-hybrid system)
- Split TEV (Tobacco etch virus protease) [4]
- Luciferase[5]
- Ubiquitin[6]
- GFP (split-GFP), e.g. EGFP (enhanced green fluorescent protein)[7][8][9]
- LacZ (beta-galactosidase)[10]
External links
- Protein–Fragment Complementation Assays: A General Strategy for the in vivo Detection of Protein-Protein Interactions – a study in the Journal of Biomolecular Techniques.
Notes
- ↑ Tarassov, K.; Messier, V.; Landry, C. R.; Radinovic, S.; Serna Molina, M. M. S.; Shames, I.; Malitskaya, Y.; Vogel, J.; Bussey, H.; Michnick, S. W. (2008). "An in Vivo Map of the Yeast Protein Interactome". Science 320 (5882): 1465–1470. doi:10.1126/science.1153878. PMID 18467557.
- ↑ Park, J. H.; Back, J. H.; Hahm, S. H.; Shim, H. Y.; Park, M. J.; Ko, S. I.; Han, Y. S. (2007). "Bacterial beta-lactamase fragmentation complementation strategy can be used as a method for identifying interacting protein pairs". Journal of microbiology and biotechnology 17 (10): 1607–1615. PMID 18156775.
- ↑ Remy, I.; Ghaddar, G.; Michnick, S. W. (2007). "Using the β-lactamase protein-fragment complementation assay to probe dynamic protein–protein interactions". Nature Protocols 2 (9): 2302–2306. doi:10.1038/nprot.2007.356. PMID 17853887.
- ↑ Wehr, M. C.; Laage, R.; Bolz, U.; Fischer, T. M.; Grünewald, S.; Scheek, S.; Bach, A.; Nave, K. A.; Rossner, M. J. (2006). "Monitoring regulated protein-protein interactions using split TEV". Nature Methods 3 (12): 985–993. doi:10.1038/nmeth967. PMID 17072307.
- ↑ Cassonnet, P.; Rolloy, C.; Neveu, G.; Vidalain, P. O.; Chantier, T.; Pellet, J.; Jones, L.; Muller, M.; Demeret, C.; Gaud, G.; Vuillier, F. O.; Lotteau, V.; Tangy, F. D.; Favre, M.; Jacob, Y. (2011). "Benchmarking a luciferase complementation assay for detecting protein complexes". Nature Methods 8 (12): 990–992. doi:10.1038/nmeth.1773. PMID 22127214.
- ↑ Dünkler, A.; Müller, J.; Johnsson, N. (2012). Detecting Protein–Protein Interactions with the Split-Ubiquitin Sensor. "Gene Regulatory Networks". Methods in molecular biology (Clifton, N.J.). Methods in Molecular Biology 786: 115–130. doi:10.1007/978-1-61779-292-2_7. ISBN 978-1-61779-291-5. PMID 21938623.
- ↑ Barnard, E.; Timson, D. J. (2010). Split-EGFP Screens for the Detection and Localisation of Protein–Protein Interactions in Living Yeast Cells. "Molecular and Cell Biology Methods for Fungi". Methods in molecular biology (Clifton, N.J.). Methods in Molecular Biology 638: 303–317. doi:10.1007/978-1-60761-611-5_23. ISBN 978-1-60761-610-8. PMID 20238279.
- ↑ Blakeley, B. D.; Chapman, A. M.; McNaughton, B. R. (2012). "Split-superpositive GFP reassembly is a fast, efficient, and robust method for detecting protein–protein interactions in vivo". Molecular BioSystems 8 (8): 2036–2040. doi:10.1039/c2mb25130b. PMID 22692102.
- ↑ Cabantous, S. P.; Nguyen, H. B.; Pedelacq, J. D.; Koraïchi, F.; Chaudhary, A.; Ganguly, K.; Lockard, M. A.; Favre, G.; Terwilliger, T. C.; Waldo, G. S. (2013). "A New Protein-Protein Interaction Sensor Based on Tripartite Split-GFP Association". Scientific Reports 3: 2854. doi:10.1038/srep02854. PMC 3790201. PMID 24092409.
- ↑ Rossi, F.; Charlton, C. A.; Blau, H. M. (1997). "Monitoring protein-protein interactions in intact eukaryotic cells by beta-galactosidase complementation". Proceedings of the National Academy of Sciences of the United States of America 94 (16): 8405–8410. PMC 22934. PMID 9237989.