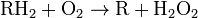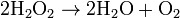Peroxisome


Peroxisomes IPA: [pɛɜˈɹɒksɪˌsoʊmz][1] (also called microbodies) are organelles found in virtually all eukaryotic cells.[2] They are involved in the catabolism of very long chain fatty acids, branched chain fatty acids, D-amino acids, and polyamines, and biosynthesis of plasmalogens, i.e. ether phospholipids critical for the normal function of mammalian brains and lungs.[3] They also contain approximately 10% of the total activity of two enzymes in the pentose phosphate pathway, which is important for energy metabolism.[3] It is vigorously debated if peroxisomes are involved in isoprenoid and cholesterol synthesis in animals.[3] Other known peroxisomal functions include the glyoxylate cycle in germinating seeds ("glyoxysomes"), photorespiration in leaves,[4] glycolysis in trypanosomes ("glycosomes"), and methanol and/or amine oxidation and assimilation in some yeasts.
Peroxisomes were identified as organelles by the Belgian cytologist Christian de Duve in 1967[5] after they had been first described by a Swedish doctoral student, J. Rhodin in 1954.[6]
Metabolic functions
A major function of the peroxisome is the breakdown of very long chain fatty acids through beta-oxidation. In animal cells, the very long fatty acids are converted to medium chain fatty acids, which are subsequently shuttled to mitochondria where they are eventually broken down to carbon dioxide and water. In yeast and plant cells, this process is exclusive for the peroxisomes.[7]
The first reactions in the formation of plasmalogen in animal cells also occur in peroxisomes. Plasmalogen is the most abundant phospholipid in myelin. Deficiency of plasmalogens causes profound abnormalities in the myelination of nerve cells, which is one reason why many peroxisomal disorders affect the nervous system.[7] Peroxisomes also play a role in the production of bile acids important for the absorption of fats and fat-soluble vitamins, such as vitamins A and K. Skin disorders are features of genetic disorders affecting peroxisome function as a result.
Peroxisomes contain oxidative enzymes, such as catalase, D-amino acid oxidase, and uric acid oxidase.[8] However the last enzyme is absent in humans, explaining the disease known as gout, caused by the accumulation of uric acid. Certain enzymes within the peroxisome, by using molecular oxygen, remove hydrogen atoms from specific organic substrates (labeled as R), in an oxidative reaction, producing hydrogen peroxide (H2O2, itself toxic):
Catalase, another peroxisomal enzyme, uses this H2O2 to oxidize other substrates, including phenols, formic acid, formaldehyde, and alcohol, by means of the peroxidation reaction:
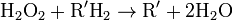 , thus eliminating the poisonous hydrogen peroxide in the process.
, thus eliminating the poisonous hydrogen peroxide in the process.
This reaction is important in liver and kidney cells, where the peroxisomes detoxify various toxic substances that enter the blood. About 25% of the ethanol humans drink is oxidized to acetaldehyde in this way.[7] In addition, when excess H2O2 accumulates in the cell, catalase converts it to H2O through this reaction:
In higher plants, peroxisomes contain also a complex battery of antioxidative enzymes such as superoxide dismutase, the components of the ascorbate-glutathione cycle, and the NADP-dehydrogenases of the pentose-phosphate pathway. It has been demonstrated that peroxisomes generate superoxide (O2•-) and nitric oxide (•NO) radicals.[9][10]
The peroxisome of plant cells is polarised when fighting fungal penetration. Infection causes a glucosinolate molecule to play an antifungal role to be made and delivered to the outside of the cell through the action of the peroxisomal proteins (PEN2 and PEN3).[11]
Peroxisome assembly
Peroxisomes can be derived from the endoplasmic reticulum and replicate by fission.[12] Peroxisome matrix proteins are translated in the cytoplasm prior to import. Specific amino acid sequences (PTS or peroxisomal targeting signal) at the C-terminus (PTS1) or N-terminus (PTS2) of peroxisomal matrix proteins signals them to be imported into the organelle. There are at least 32 known peroxisomal proteins, called peroxins,[13] which participate in the process of peroxisome assembly. Proteins do not have to unfold to be imported into the peroxisome. The protein receptors, the peroxins PEX5 and PEX7, accompany their cargoes (containing a PTS1 or a PTS2 amino acid sequence, respectively) all the way into the peroxisome where they release the cargo and then return to the cytosol - a step named recycling. A model describing the import cycle is referred to as the extended shuttle mechanism.[14] There is now evidence that ATP hydrolysis is required for the recycling of receptors to the cytosol. Also, ubiquitination appears to be crucial for the export of PEX5 from the peroxisome, to the cytosol.
Associated medical conditions
Peroxisomal disorders are a class of medical conditions that typically affect the human nervous system as well as many other organ systems. Two common examples are X-linked adrenoleukodystrophy and peroxisome biogenesis disorders.[15][16]
Genes
PEX genes encode the protein machinery ("peroxins") required for proper peroxisome assembly, as described above. Membrane assembly and maintenance requires three of these (peroxins 3, 16, and 19) and may occur without the import of the matrix (lumen) enzymes. Proliferation of the organelle is regulated by Pex11p.
Genes that encode peroxin proteins include: PEX1, PEX2 - PXMP3, PEX3, PEX5, PEX6, PEX7, PEX10, PEX11A, PEX11B, PEX11G, PEX12, PEX13, PEX14, PEX16, PEX19, PEX26, PEX28, PEX30, and PEX31.
Evolutionary origins
The protein content of peroxisomes varies across species, but the presence of proteins common to many species has been used to suggest an endosymbiotic origin; that is, peroxisomes evolved from bacteria that invaded larger cells as parasites, and very gradually evolved a symbiotic relationship.[17] However, this view has been challenged by recent discoveries.[18] For example, peroxisome-less mutants can restore peroxisomes upon introduction of the wild-type gene.
Two independent evolutionary analyses of the peroxisomal proteome found homologies between the peroxisomal import machinery and the ERAD pathway in the endoplasmic reticulum,[19][20] along with a number of metabolic enzymes that were likely recruited from the mitochondria.[20] Recently, it has been suggested that the peroxisome may have had an actinobacterial origin,[21] however, this is controversial.[22]
Other related organelles
Other organelles of the microbody family related to peroxisomes include glyoxysomes of plants and filamentous fungi, glycosomes of kinetoplastids[23] and Woronin bodies of filamentous fungi.
References
- ↑ "Peroxisome". Online DIctionary. Merriam-Webster. Retrieved 19 June 2013.
- ↑ Gabaldón T (2010). "Peroxisome diversity and evolution". Philos Trans R Soc Lond B Biol Sci. 365 (1541): 765–73. doi:10.1098/rstb.2009.0240. PMC 2817229. PMID 20124343.
- ↑ 3.0 3.1 3.2 Wanders RJ, Waterham HR (2006). "Biochemistry of mammalian peroxisomes revisited". Annu. Rev. Biochem. 75: 295–332. doi:10.1146/annurev.biochem.74.082803.133329. PMID 16756494.
- ↑ Evert, R.F.; Eichhorn, S.E. (2006). Esau's Plant Anatomy: Meristems, Cells, and Tissues of the Plant Body: Their Structure, Function, and Development. John Wiley & Sons. ISBN 9780471738435.
- ↑ de Duve C (1969). "The peroxisome: a new cytoplasmic organelle". Proc. R. Soc. Lond., B, Biol. Sci. 173 (30): 71–83. doi:10.1098/rspb.1969.0039. PMID 4389648.
- ↑ Rhodin, J (1954). "Correlation of ultrastructural organization and function in normal and experimentally changed proximal tubule cells of the mouse kidney". Doctorate Thesis. Karolinska Institutet, Stockholm.
- ↑ 7.0 7.1 7.2 Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002). "Chapter 12: Peroxisomes". Molecular Biology of the Cell (Fourth ed.). New York: Garland Science. ISBN 0-8153-3218-1.
- ↑ del Río LA, Sandalio LM, Palma JM, Bueno P, Corpas FJ (1992). "Metabolism of oxygen radicals in peroxisomes and cellular implications". Free Radic. Biol. Med. 13 (5): 557–80. doi:10.1016/0891-5849(92)90150-F. PMID 1334030.
- ↑ Corpas FJ, Barroso JB, del Río LA (April 2001). "Peroxisomes as a source of reactive oxygen species and nitric oxide signal molecules in plant cells". Trends Plant Sci. 6 (4): 145–50. doi:10.1016/S1360-1385(01)01898-2. PMID 11286918.
- ↑ Corpas FJ, Barroso JB, Carreras A, Quirós M, León AM, Romero-Puertas MC, Esteban FJ, Valderrama R, Palma JM, Sandalio LM, Gómez M, del Río LA (September 2004). "Cellular and subcellular localization of endogenous nitric oxide in young and senescent pea plants". Plant Physiol. 136 (1): 2722–33. doi:10.1104/pp.104.042812. PMC 523336. PMID 15347796.
- ↑ Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, Humphry M, Consonni C, Panstruga R, Sanchez-Vallet A, Molina A, Schulze-Lefert P (January 2009). "A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense". Science 323 (5910): 101–6. doi:10.1126/science.1163732. PMID 19095900.
- ↑ Hoepfner D, Schildknegt D, Braakman I, Philippsen P, Tabak HF (2005). "Contribution of the endoplasmic reticulum to peroxisome formation". Cell 122 (1): 85–95. doi:10.1016/j.cell.2005.04.025. PMID 16009135.
- ↑ Saleem RA, Smith JJ, Aitchison JD (2006). "Proteomics of the Peroxisome". Biochim. Biophys. Acta 1763 (12): 1541–51. doi:10.1016/j.bbamcr.2006.09.005. PMC 1858641. PMID 17050007.
- ↑ Dammai V, Subramani S (April 2001). "The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisomal matrix and recycled to the cytosol". Cell 105 (2): 187–96. PMID 11336669.
- ↑ Depreter M, Espeel M, Roels F (June 2003). "Human peroxisomal disorders". Microsc. Res. Tech. 61 (2): 203–23. doi:10.1002/jemt.10330. PMID 12740827.
- ↑ Roels F, De Bie S, Schutgens RBH, Besley GTN (1995). "Diagnostic laboratory methods in peroxisomal disorders". Journal of Inherited Metabolic Disease 18 (Supplement 1): 1–229.
- ↑ Lazarow PB, Fujiki Y (1985). "Biogenesis of peroxisomes". Annu. Rev. Cell Biol. 1: 489–530. doi:10.1146/annurev.cb.01.110185.002421. PMID 3916321.
- ↑ Fagarasanu A, Fagarasanu M, Rachubinski, RA (2007). "Maintaining peroxisome populations: a story of division and inheritance". Annu. Rev. Cell Dev. Biol. 23: 321–344. doi:10.1146/annurev.cellbio.23.090506.123456. PMID 17506702.
- ↑ Schlüter A, Fourcade S, Ripp R, Mandel JL, Poch O, Pujol A (2006). "The evolutionary origin of peroxisomes: an ER-peroxisome connection". Mol Biol Evol 23 (4): 838–45. doi:10.1093/molbev/msj103. PMID 16452116.
- ↑ 20.0 20.1 Gabaldón T, Snel B, van Zimmeren F, Hemrika W, Tabak H, Huynen MA (2006). "Origin and evolution of the peroxisomal proteome". Biol. Direct 1: 8. doi:10.1186/1745-6150-1-8. PMC 1472686. PMID 16556314.
- ↑ Duhita, et al; Le, HA; Satoshi, S; Kazuo, H; Daisuke, M; Takao, S (2009). "The origin of peroxisomes: The possibility of an actinobacterial symbiosis". Gene 450 (1–2): 18–24. doi:10.1016/j.gene.2009.09.014. PMID 19818387.
- ↑ Gabaldón T, Capella-Gutiérrez S (October 2010). "Lack of phylogenetic support for a supposed actinobacterial origin of peroxisomes". Gene 465 (1–2): 61–5. doi:10.1016/j.gene.2010.06.004. PMID 20600706.
- ↑ Blattner J, Swinkels B, Dörsam H, Prospero T, Subramani S, Clayton C (December 1992). "Glycosome assembly in trypanosomes: variations in the acceptable degeneracy of a COOH-terminal microbody targeting signal". J. Cell Biol. 119 (5): 1129–36. doi:10.1083/jcb.119.5.1129. PMC 2289717. PMID 1447292.
Further reading
- Mateos RM, León AM, Sandalio LM, Gómez M, del Río LA, Palma JM (December 2003). "Peroxisomes from pepper fruits (Capsicum annuum L.): purification, characterisation and antioxidant activity". J. Plant Physiol. 160 (12): 1507–16. doi:10.1078/0176-1617-01008. PMID 14717445.
External links
| Wikiversity has learning materials about Peroxisomes at |
| Wikimedia Commons has media related to Peroxisomes. |
![]() This article incorporates public domain material from the NCBI document "Science Primer".
This article incorporates text from the public domain Pfam and InterPro IPR006708
This article incorporates public domain material from the NCBI document "Science Primer".
This article incorporates text from the public domain Pfam and InterPro IPR006708
| ||||||||||||||||||||
| |||||||||||||||||