Organ-on-a-chip
An Organ-on-a-Chip (OC) is a multi-channel 3-D microfluidic cell culture chip that simulates the activities, mechanics and physiological response of entire organs and organ systems.[1] It constitutes the subject matter of significant biomedical engineering research, more precisely in bio-MEMS. The convergence of Lab-on-Chips (LOCs) and cell biology has permitted the study of human physiology in an organ-specific context, introducing a novel model of in vitro multicellular human organisms. One day, they will perhaps abolish the need for animals in drug development and toxin testing.
Although multiple publications claim to have translated organ functions onto this interface, the movement towards this microfluidic application is still in its infancy. Organs-on-chips will vary in design and approach between different researchers. As such, validation and optimization of these systems will likely be a long process. Organs that have been simulated by microfluidic devices include the heart, the lung, kidney, artery, bone, cartilage, skin and more.
Nevertheless, building valid artificial organs requires not only a precise cellular manipulation, but a detailed understanding of the human body’s fundamental intricate response to any event. A common concern with Organs-on-Chips lies in the isolation of organs during testing. “If you don’t use as close to the total physiological system that you can, you’re likely to run into troubles”[1] says William Haseltine, founder of Rockville, MD. Microfabrication, microelectronics and microfluidics offer the prospect of modeling sophisticated in vitro physiological responses under accurately simulated conditions.
Brief Overview of Lab-on-Chips (LOCs)
A Lab-on-a-Chip is a device that integrates one or several laboratory functions on a single chip that deals with handling particles in hollow microfluidic channels. It has been developed for over a decade. Advantages in handling particles at such a small scale include lowering fluid volume consumption (lower reagents costs, less waste), increasing portability of the devices, increasing process control (due to quicker thermo-chemical reactions) and decreasing fabrication costs. Additionally, microfluidic flow is entirely laminar (i.e., no turbulence). Consequently, there is virtually no mixing between neighboring streams in one hollow channel. In cellular biology convergence, this rare property in fluids has been leveraged to better study complex cell behaviors, such as cell motility in response to chemotactic stimuli, stem cell differentiation, axon guidance, subcellular propagation of biochemical signaling and embryonic development.[2]
Transitioning from 3D Cell-Culture Models to Organs-on-Chips
3D cell-culture models exceed 2D culture systems by promoting higher levels of cell differentiation and tissue organization. 3D culture systems are more successful because the flexibility of the ECM gels accommodates shape changes and cell-cell connections – formerly prohibited by rigid 2D culture substrates. Nevertheless, even the best 3D culture models fail to mimic an organ’s cellular properties in many aspects,[2] including tissue-to-tissue interfaces (e.g., epithelium and vascular endothelium), spatiotemporal gradients of chemicals, and the mechanically active microenvironments (e.g. arteries’ vasoconstriction and vasodilator responses to temperature differentials). The application of microfluidics in Organs-on-Chips enables the efficient transport and distribution of nutrients and other soluble cues throughout the viable 3D tissue constructs. Organs-on-Chips are referred to as the next wave of 3D cell-culture models that mimic whole living organs’ biological activities, dynamic mechanical properties and biochemical functionalities.[1]
Organs
Lung-on-a-Chip
Lung-on-a-chips are being designed in an effort to improve the physiological relevance of existing in vitro alveolar-capillary interface models.[3] Such a multifunctional microdevice can reproduce key structural, functional and mechanical properties of the human alveolar-capillary interface (i.e., the fundamental functional unit of the living lung).
- Example
- Dongeun Huh from Wyss Institute for Biologically Inspired Engineering at Harvard describes their fabrication of a system containing two closely apposed microchannels separated by a thin (10µm) porous flexible membrane made of PDMS.[4] The device largely comprises three microfluidic channels, and only the middle one holds the porous membrane. Culture cells were grown on either side of the membrane: human alveolar epithelial cells on one side, and human pulmonary microvascular endothelial cells on the other.
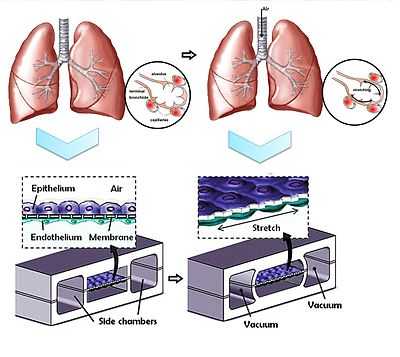
- The compartmentalization of the channels facilitates not only the flow of air as a fluid which delivers cells and nutrients to the apical surface of the epithelium, but also allows for pressure differences to exist between the middle and side channels. During normal inspiration in a human’s respiratory cycle, intrapleural pressure decreases, triggering an expansion of the alveoli. As air is pulled into the lungs, alveolar epithelium and the coupled endothelium in the capillaries are stretched. Since a vacuum is connected to the side channels, a decrease in pressure will cause the middle channel to expand, thus stretching the porous membrane and subsequently, the entire alveolar-capillary interface. The pressure-driven dynamic motion behind the stretching of the membrane, also described as a cyclic mechanical strain (valued at approximately 10%), significantly increases the rate of nanoparticle translocation across the porous membrane, when compared to a static version of this device, and to a Transwell culture system.
- In order to fully validate the biological accuracy of a device, its whole-organ responses must be evaluated. In this instance, researchers inflicted injuries to the cells:
- Pulmonary inflammation
- Pulmonary inflammatory responses entail a multistep strategy, but alongside an increased production of epithelial cells and an early response release of cytokines, the interface should undergo an increased number of leukocyte adhesion molecules.[5] In Huh’s experiment, the pulmonary inflammation was simulated by introducing medium containing a potent proinflammatory mediator. Only hours after the injury was caused, the cells in the microfluidic device subjected to a cyclic strain reacted in accordance with the previously mentioned biological response.
- Pulmonary infection
- Living E-coli bacteria was used to demonstrate how the system can even mimic the innate cellular response to a bacterial pulmonary infection. The bacteria were introduced onto the apical surface of the alveolar epithelium. Within hours, neutrophils were detected in the alveolar compartment, meaning they had transmigrated from the vascular microchannel where the porous membrane had phagocytized the bacteria.
Additionally, researchers believe the potential value of this lung-on-a-chip system will aid in toxicology applications. By investigating the pulmonary response to nanoparticles, researchers hope to learn more about health risks in certain environments, and correct previously oversimplified in vitro models. Because a microfluidic lung-on-a-chip can more exactly reproduce the mechanical properties of a living human lung, its physiological responses will be quicker and more accurate than a Transwell culture system. Nevertheless, published studies admit that responses of a lung-on-a-chip don’t yet fully reproduce the responses of native alveolar epithelial cells.
Heart-on-a-Chip
Past efforts to replicate in vivo cardiac tissue environments have proven to be challenging due to difficulties when mimicking contractility and electrophysiological responses. Such features would greatly increase the accuracy of in vitro experiments.
Microfluidics has already contributed to in vitro experiments on cardiomyocytes, which generate the electrical impulses that control the heart rate.[6] For instance, researchers have built an array of PDMS microchambers, aligned with sensors and stimulating electrodes as a tool that will electrochemically and optically monitor the cardiomyocytes’ metabolism.[7] Another lab-on-a-chip similarly combined a microfluidic network in PDMS with planar microelectrodes, this time to measure extracellular potentials from single adult murine cardiomyocytes.[8]
- Example

- A reported design of a heart-on-a-chip claims to have built “an efficient means of measuring structure-function relationships in constructs that replicate the hierarchical tissue architectures of laminar cardiac muscle.”[9] This chip determines that the alignment of the myocytes in the contractile apparatus made of cardiac tissue and the gene expression profile (affected by shape and cell structure deformation) contributes to the force produced in cardiac contractility. This heart-on-a-chip is a biohybrid construct: an engineered anisotropic ventricular myocardium is an elastomeric thin film.
- The design and fabrication process of this particular microfluidic device entails first covering the edges of a glass surface with tape (or any protective film) such as to contour the substrate’s desired shape. A spin coat layer of PNIPA is then applied. After its dissolution, the protective film is peeled away, resulting in a self-standing body of PNIPA. The final steps involve the spin coating of protective surface of PDMS over the cover slip and curing. Muscular thin films (MTF) enable cardiac muscle monolayers to be engineered on a thin flexible substrate of PDMS.[10] In order to properly seed the 2D cell culture, a microcontact printing technique was used to lay out a fibronectin “brick wall” pattern on the PDMS surface. Once the ventricular myocytes were seeded on the functionalized substrate, the fibronectin pattern oriented them to generate an anisotropic monolayer.
- After the cutting of the thin films into two rows with rectangular teeth, and subsequent placement of the whole device in a bath, electrodes stimulate the contraction of the myocytes via a field-stimulation – thus curving the strips/teeth in the MTF. Researchers have developed a correlation between tissue stress and the radius of curvature of the MTF strips during the contractile cycle, validating the demonstrated chip as a “platform for quantification of stress, electrophysiology and cellular architecture.”[9]
Kidney-on-a-Chip
Renal cells and nephrons have already been simulated by microfluidic devices. “Such cell cultures can lead to new insights into cell and organ function and be used for drug screening”.[11] A kidney-on-a-chip device has the potential to accelerate research encompassing artificial replacement for lost kidney function. Nowadays, dialysis requires patients to go to a clinic up to three times per week. A more transportable and accessible form of treatment would not only increase the patient’s overall health (by increasing frequency of treatment), but the whole process would become more efficient and tolerable.[12] Artificial kidney research is striving to bring transportability, wearability and perhaps implantation capability to the devices through innovative disciplines: microfluidics, miniaturization and nanotechnology.[13]
- Example – Nephron-on-a-Chip
The nephron is the functional unit of the kidney and is composed of a glomerulus and a tubular component.[14] Researchers at MIT claim to have designed a bioartificial device that replicates the function of the nephron’s glomerulus, proximal convoluted tubule and loop of Henle.
Each part of the device has its unique design, generally consisting of two microfabricated layers separated by a membrane. The only inlet to the microfluidic device is designed for the entering blood sample. In the glomerulus’ section of the nephron, the membrane allows certain blood particles through its wall of capillary cells, composed by the endothelium, basement membrane and the epithelial podocytes. The fluid that is filtered from the capillary blood into Bowman’s space is called filtrate or primary urine.[15]

In the tubules, some substances are added to the filtrate as part of the urine formation, and some substances reabsorbed out of the filtrate and back into the blood. The first segment of these tubules is the proximal convoluted tubule. This is where the almost complete absorption of nutritionally important substances takes place. In the device, this section is merely a straight channel, but blood particles going to the filtrate have to cross the previously mentioned membrane and a layer of renal proximal tubule cells. The second segment of the tubules is the loop of Henle where the reabsorption of water and ions from the urine takes place. The device’s looping channels strives to simulate the countercurrent mechanism of the loop of Henle. Likewise, the loop of Henle requires a number of different cell types because each cell type has distinct transport properties and characteristics. These include the descending limb cells, thin ascending limb cells, thick ascending limb cells, cortical collecting duct cells and medullary collecting duct cells.[14]
One step towards validating the microfluidic device’s simulation of the full filtration and reabsorption behavior of a physiological nephron would include demonstrating that the transport properties between blood and filtrate are identical with regards to where they occur and what is being let in by the membrane. For example, the large majority of passive transport of water occurs in the proximal tubule and the descending thin limb, or the active transport of NaCl largely occurs in the proximal tubule and the thick ascending limb. The device’s design requirements would require the filtration fraction in the glomerulus to vary between 15%-20%, or the filtration reabsorption in the proximal convoluted tubule to vary between 65%-70%, and finally the urea concentration in urine (collected at one of the two outlets of the device) to vary between 200-400mM.[16]
Artery-on-a-Chip
Cardiovascular diseases are often caused by changes in structure and function of small blood vessels. For instance, self-reported rates of hypertension suggest that the rate is increasing, says a 2003 report from the National Health and Nutrition Examination Survey.[17] A microfluidic platform simulating the biological response of an artery could not only enable organ-based screens to occur more frequently throughout a drug development trial, but also yield a comprehensive understanding of the underlying mechanisms behind pathologic changes in small arteries and develop better treatment strategies. Axel Gunther from the University of Toronto argues that such MEMS-based devices could potentially help in the assessment of a patient’s microvascular status in a clinical setting (personalized medicine).[18]
Conventional methods used to examine intrinsic properties of isolated resistance vessels (arterioles and small arteries with diameters varying between 30 µm and 300 µm) include the pressure myography technique. However, such methods currently require manually skilled personnel and are not scalable. An artery-on-a-chip could overcome several of these limitations by accommodating an artery onto a platform which would be scalable, inexpensive and possibly automated in its manufacturing.
- Example
An organ-based microfluidic platform has been developed as a lab-on-a-chip onto which a fragile blood vessel can be fixed, allowing for determinants of resistance artery malfunctions to be studied.
The artery microenvironment is characterized by surrounding temperature, transmural pressure, and luminal & abluminal drug concentrations. The multiple inputs from a microenvironment cause a wide range of mechanical or chemical stimuli on the smooth muscle cells (SMCs) and endothelial cells (ECs) that line the vessel’s outer and luminal walls, respectively. Endothelial cells are responsible for releasing vasoconstriction and vasodilator factors, thus modifying tone. Vascular tone is defined as the degree of constriction inside a blood vessel relative to its maximum diameter.[19] Pathogenic concepts currently believe that subtle changes to this microenvironment have pronounced effects on arterial tone and can severely alter peripheral vascular resistance. The engineers behind this design believe that a specific strength lies in its ability to control and simulate heterogeneous spatiotemporal influences found within the microenvironment, whereas myography protocols have, by virtue of their design, only established homogeneous microenvironments.[18] They proved that by delivering phenylephrine through only one of the two channels providing superfusion to the outer walls, the drug-facing side constricted much more than the drug opposing side.

The artery-on-a-chip is designed for reversible implantation of the sample. The device contains a microchannel network, an artery loading area and a separate artery inspection area. There is a microchannel used for loading the artery segment, and when the loading well is sealed, it is also used as a perfusion channel, to replicate the process of nutritive delivery of arterial blood to a capillary bed in the biological tissue.[20] Another pair of microchannels serves to fix the two ends of the arterial segment. Finally, the last pair of microchannels is used to provide superfusion flow rates, in order to maintain the physiological and metabolic activity of the organ by delivering a constant sustaining medium over the abluminal wall. A thermoelectric heater and a thermoresistor are connected to the chip and maintain physiological temperatures at the artery inspection area.
The protocol of loading and securing the tissue sample into the inspection zone helps understand how this approach acknowledges whole organ functions. After immersing the tissue segment into the loading well, the loading process is driven by a syringe withdrawing a constant flow rate of buffer solution at the far end of the loading channel. This causes the transport of the artery towards its dedicated position. This is done with closed fixation and superfusion in/outlet lines. After stopping the pump, sub-atmospheric pressure is applied through one of the fixation channels. Then after sealing the loading well shut, the second fixation channel is subjected to a sub-atmospheric pressure. Now the artery is symmetrically established in the inspection area, and a transmural pressure is felt by the segment. The remaining channels are opened and constant perfusion and superfusion are adjusted using separate syringe pumps.[18]
Human-on-a-Chip
Researchers are working towards building a multi-channel 3D microfluidic cell culture system that compartmentalizes microenvironments in which 3D cellular aggregates are cultured to mimic multiple organs in the body.[21] Most organ-on-a-chip models today only culture one cell type, so even though they may be valid models for studying whole organ functions, the systemic effect of a drug on the human body is not verified.

In particular, an integrated cell culture analog (µCCA) was developed and included lung cells, drug-metabolizing liver and fat cells. The cells were linked in a 2D fluidic network with culture medium circulating as a blood surrogate, thus efficiently providing a nutritional delivery transport system, while simultaneously removing wastes from the cells.[22] “The development of the µCCA laid the foundation for a realistic in vitro pharmacokinetic model and provided an integrated biomimetic system for culturing multiple cell types with high fidelity to in vivo situations”, claim C. Zhang et al. They have developed a microfluidic human-on-a-chip, culturing four different cell types to mimic four human organs: liver, lung, kidney and fat.[23] They focused on developing a standard serum-free culture media that would be valuable to all cell types included in the device. Optimized standard media are generally targeted to one specific cell-type, whereas a human-on-a-chip will evidently require a common medium (CM). In fact, they claim to have identified a cell culture CM that, when used to perfuse all cell cultures in the microfluidic device, maintains the cells’ functional levels. Heightening the sensitivity of the in vitro cultured cells ensures the validity of the device, or that any drug injected into the microchannels will stimulate an identical physiological and metabolic reaction from the sample cells as whole organs in humans.
With more extensive development of this kind of chip, pharmaceutical companies will potentially be able to measure direct effects of one organ’s reaction on another. For instance, the delivery of biochemical substances would be screened to confirm that even though it may benefit one cell type, it does not compromise the functions of others. It is probably already possible to print these organs with 3D printers,[24] but the cost is too high. Designing whole body biomimetic devices addresses a major reservation that pharmaceutical companies have towards organs-on-chips, namely the isolation of organs. As these devices become more and more accessible, the complexity of the design increases exponentially. Systems will soon have to simultaneously provide mechanical perturbation and fluid flow through a circulatory system. “Anything that requires dynamic control rather than just static control is a challenge”, says Takayama from the University of Michigan.[25]
Replacing Animal Testing with Organs-on-Chips
In the early phase of drug development, animal models were the only way of obtaining in vivo data that would predict the human pharmacokinetic responses. However, experiments on animals are lengthy, expensive and controversial. For example, animal models are often subjected to mechanical or chemical techniques that simulate human injuries. There are also concerns with regards to the validity of such animal models, due to deficiency in cross-species extrapolation.[26] Moreover, animal models offer very limited control of individual variables and can be cumbersome to harvest specific information.
Therefore, mimicking a human’s physiological responses in an in vitro model needs to be made more affordable, and needs to offer cellular level control in biological experiments: biomimetic microfluidic systems could replace animal testing. The development of MEMS-based biochips that reproduce complex organ-level pathological responses could revolutionize many fields, including toxicology and the developmental process of pharmaceuticals and cosmetics that rely on animal testing and clinical trials.[27]
References
- ↑ 1.0 1.1 1.2 Melinda Wenner Moyer , “Organs-on-a-Chip for Faster Drug Development”, Scientific American 25 Feb. 2011
- ↑ 2.0 2.1 Dongeun Huh, Geraldine A. Hamilton and Donald E. Ingber (2011), From 3D cell culture to organs-on-chips
- ↑ Diviya D. Nalayanda, Christopher Puleo, William B. Fulton, Leilani M. Sharpe, Tza-Huei Wang, Fizan Abdullah (2009), “An open-access microfluidic model for lung-specific functional studies at an air-liquid interface”
- ↑ D. Huh, B. D. Matthews, A. Mammoto, M. Montoya-Zavala, H. Y. Hsin, D. E. Ingber (2010), “Reconstituting Organ-Level Lung Functions on a Chip”
- ↑ M.I. Hermanns et al., Cell Tissue Res. 336, 91 (2009)
- ↑ Franke, W. W., Borrmann, C. M., Grund, C. and Pieperhoff, S. (2006). The area composite of adhering junctions connecting heart muscle cells of vertebrates: Molecular definition in intercalated disks of cardiomyocytes by immunoelectron microscopy of desmosomal proteins. Eur J Cell Biol 85, 69-82.
- ↑ W. Cheng, N. Klauke, H. Sedgwick, G. L. Smith and J. M. Cooper, Lab on a Chip, 2006, 6, 1424-1431
- ↑ A. Werdich, E. A. Lima, B. Ivanov, I. Ges, M. E. Anderson, J. P. Wikswo and F. J. Baudenbacher, Lab on a Chip, 2004, 4, 357–362
- ↑ 9.0 9.1 Anna Grosberg, Patrick W. Alford, Megan L. McCain and Kevin Kit Parker (2011), “Ensembles of engineered cardiac tissues for physiological and pharmacological study: Heart on a chip”
- ↑ Patrick W. Alford, Adam W. Feinberg, Sean P. Sheehy, Kevin K. Parker (2009). Biohybrid thin films for measuring contractility in engineered cardiovascular muscle
- ↑ Kidney on a Chip, Highlights in Chemical Biology, RSC Publishing
- ↑ Cruz D, Bellomo R, Kellum JA, De Cal M, Ronco C. The future of extracorporeal support. Crit Care Med 2008;36(4 Suppl):S243-52.
- ↑ C. Ronco, A. Davenport, V. Gura (2011), The future of the artificial kidney: moving towards wearable and miniaturized devices
- ↑ 14.0 14.1 Maton, Anthea; Jean Hopkins, Charles William McLaughlin, Susan Johnson, Maryanna Quon Warner, David LaHart, Jill D. Wright (1993). Human Biology and Health. Englewood Cliffs, New Jersey, USA
- ↑ Koeppen B, Stanton B. Renal physiology; 3rd ed. St Louis, MO: Mosby, 2001
- ↑ E. Weinberg, M. Kaazempur-Mofrad, J. Borenstein (2008), “Concept and computational design for a bioartificial nephron-on-a-chip”
- ↑ Ihab Hajjar, MD, MS (2003), Trends in Prevalence, Awareness, Treatment, and Control of Hypertension in the United States, 1988-2000
- ↑ 18.0 18.1 18.2 A. Gunther, S. Yasotharan, A. Vagaon, C. Lochovsky, S. Pinto, J. Yang, C. Lau, J. Voigtlaender-Bolz, S. Bolz (2010), “A microfluidic platform for probing small artery structure and function”
- ↑ Lippincott, Williams & Wilkins (2011), Cardiovascular Physiology Concepts 2nd Ed.
- ↑ N. Marieb, K. Hoehn (2006), Human Anatomy & Physiology 7th Ed.
- ↑ C. Luni, E. Serena, N. Elvassore (2014), “Human-on-chip for therapy development and fundamental science”, Curr Opin Biotech ‘’’25’’’, 45-50
- ↑ K. Viravaidya and M. L. Shuler (2004), Biotechnol. Prog., ‘’’20’’’, 590-597
- ↑ C. Zhang, Z. Zhao, N. Rahim, D. Noort, H. Yu (2009), “Towards a human-on-chip: Culturing multiple cell types on a chip with compartmentalized microenvironments”
- ↑ 3D Printing Organs
- ↑ Monya Baker (2011), “Tissue models: A living system on a chip”, Nature ‘’’471’’’, 661-665
- ↑ I. Roberts & al. (2002), “Does animal experimentation inform human healthcare? Observations form a systematic review of international animal experiments on fluid resuscitation”
- ↑ Anja van de Stolpe and Jaap den Toonder, "Workshop meeting report Organs-on-Chips: human disease models", RSC Publishing (2013)
External links
| Wikiversity has learning materials about Ethical medical research/Alternatives to animal testing#Biochip |