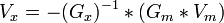Metabolic engineering

Metabolic engineering is the practice of optimizing genetic and regulatory processes within cells to increase the cells' production of a certain substance. These processes are chemical networks that use a series of biochemical reactions and enzymes that allow cells to convert raw materials into molecules necessary for the cell’s survival. Metabolic engineering specifically seeks to mathematically model these networks, calculate a yield of useful products, and pin point parts of the network that constrain the production of these products.[1] Genetic engineering techniques can then be used to modify the network in order to relieve these constraints. Once again this modified network can be modeled to calculate the new product yield.
The ultimate goal of metabolic engineering is to be able to use these organisms to produce valuable substances on an industrial scale in a cost effective manner. Current examples include producing beer, wine, cheese, pharmaceuticals, and other biotechnology products.
Since cells use these metabolic networks for their survival, changes can have drastic effects on the cells' ability to survive. Therefore, trade-offs in metabolic engineering arise between the cells ability to produce the desired substance and its natural survival needs. Therefore, instead of directly deleting and/or overexpressing the genes that encode for metabolic enzymes, the current focus is to target the regulatory networks in a cell to efficiently engineer the metabolism.[2]
History and applications of metabolic engineering
In the past, to increase the productivity of a desired metabolite, a microorganism was genetically modified by chemically induced mutation, and the mutant strain that overexpressed the desired metabolite was then chosen.[3] However, one of the main problem with this technique was that the metabolic pathway for the production of that metabolite was not analyzed, and as a result, constraints to production and relevant pathway enzymes to be modified were unknown.[3] In 1990s, a new technique called metabolic engineering emerged. This technique analyzes the metabolic pathway of a microorganism, and determines the constraints and their effects on the production of desired compounds. It then uses genetic engineering to relieve these constraints. Some examples of successful metabolic engineering are the following: (i) Identification of constraints to lysine production in corynebacterium glutamicum and insertion of new genes to relieve these constraints to improve production[4] (ii) Engineering of a new fatty acid biosynthesis pathway, called reversed beta oxidation pathway, that is more efficient than the native pathway in producing fatty acids and alcohols which can potentially be catalytically converted to chemicals and fuels[5] (iii) Improved production of DAHP an aromatic metabolite produced by E.coli that is an intermediate in the production of aromatic amino acids.[6] It was determined through metabolic flux analysis that the theoretical maximal yield of DAHP per glucose molecule utilized, was 3/7. This is because some of the carbon from glucose is lost as carbon dioxide, instead of being utilized to produce DAHP. Also, one of the metabolites (PEP, or phosphoenolpyruvate) that are used to produce DAHP, was being converted to pyruvate (PYR) to transport glucose into the cell, and therefore, was no longer available to produce DAHP. In order to relieve the shortage of PEP and increase yield, Patnaik et al. used genetic engineering on E.coli to introduce a reaction that converts PYR back to PEP. Thus, the PEP used to transport glucose into the cell is regenerated, and can be used to make DAHP. This resulted in a new theoretical maximal yield of 6/7 - double that of the native E.coli system.
At the industrial scale, metabolic engineering is becoming more convenient and cost effective. According to the Biotechnology Industry Organization, " more than 50 biorefinery facilities are being built across North America to apply metabolic engineering to produce biofuels and chemicals from renewable biomass which can help reduce greenhouse gas emissions ". Potential biofuels include short-chain alcohols and alkanes (to replace gasoline), fatty acid methyl esters and fatty alcohols (to replace diesel), and fatty acid-and isoprenoid-based biofuels (to replace diesel).[7]
Metabolic flux analysis
An analysis of metabolic flux can be found at Flux balance analysis
Setting up a metabolic pathway for analysis
The first step in the process is to identify a desired goal to achieve through the improvement or modification of an organism's metabolism. Reference books and online databases are used to research reactions and metabolic pathways that are able to produce this product or result. These databases contain copious genomic and chemical information including pathways for metabolism and other cellular processes. Using this research, an organism is chosen that will be used to create the desired product or result. Considerations that are taken into account when making this decision are how close the organism's metabolic pathway is to the desired pathway, the maintenance costs associated with the organism, and how easy it is to modify the pathway of the organism. Escherichia coli (E. coli) is widely used in metabolic engineering to synthesize a wide variety of products such as amino acids because it is relatively easy to maintain and modify.[8] If the organism does not contain the complete pathway for the desired product or result, then genes that produce the missing enzymes must be incorporated into the organism.
Analyzing a metabolic pathway
The completed metabolic pathway is modeled mathematically to find the theoretical yield of the product or the reaction fluxes in the cell. A flux is the rate at which a given reaction in the network occurs. Simple metabolic pathway analysis can be done by hand, but most require the use of software to perform the computations.[9] These programs use complex linear algebra algorithms to solve these models. To solve a network using the equation for determined systems shown below, one must input the necessary information about the relevant reactions and their fluxes. Information about the reaction (such as the reactants and stoichiometry) are contained in the matrices Gx and Gm. Matrices Vm and Vx contain the fluxes of the relevant reactions. When solved, the equation yields the values of all the unknown fluxes (contained in Vx).
Determining the optimal genetic manipulations
After solving for the fluxes of reactions in the network, it is necessary to determine which reactions may be altered in order to maximize the yield of the desired product. To determine what specific genetic manipulations to perform, it is necessary to use computational algorithms, such as OptGene or OptFlux.[10] They provide recommendations for which genes should be overexpressed, knocked out, or introduced in a cell to allow increased production of the desired product. For example, if a given reaction has particularly low flux and is limiting the amount of product, the software may recommend that the enzyme catalyzing this reaction should be overexpressed in the cell to increase the reaction flux. The necessary genetic manipulations can be performed using standard molecular biology techniques. Genes may be overexpressed or knocked out from an organism, depending on their effect on the pathway and the ultimate goal.[11]
Experimental measurements
In order to create a solvable model, it is often necessary to have certain fluxes already known or experimentally measured. In addition, in order to verify the effect of genetic manipulations on the metabolic network (to ensure they align with the model), it is necessary to experimentally measure the fluxes in the network. To measure reaction fluxes, carbon flux measurements are made using carbon-13 isotopic labeling.[12] The organism is fed a mixture that contains molecules where specific carbons are engineered to be carbon-13 atoms, instead of carbon-12. After these molecules are used in the network, downstream metabolites also become labeled with carbon-13, as they incorporate those atoms in their structures. The specific labeling pattern of the various metabolites is determined by the reaction fluxes in the network. Labeling patterns may be measured using techniques such as Gas chromatography-mass spectrometry (GC-MS) along with computational algorithms to determine reaction fluxes.
References
- ↑ Yang, Y.T., Bennet, G. N., San, K.Y., (1998) Genetic and Metabolic Engineering, Electronic Journal of Biotechnology, ISSN 07117-3458
- ↑ Vemuri, G.M, Aristidou, A.A, (2005) Metabolic Engineering in the -omics Era: Elucidating and Modulating Regulatory Networks, Microbial Mol Biology Review vol. 69: 197-216
- ↑ 3.0 3.1 Voit,Eberhard.,Torres,Nestor V.(2002)." Pathways Analysis and Optimization in Metabolic Engineering." Cambridge:University Press,p.ix-x
- ↑ Stephanopoulos, G. N., Aristidou, A. A., Nielsen, J. (1998). " Metabolic Engineering: Principles and Methodologies ". San Diego: Academic Press
- ↑ Dellomonaco, Clementina.(2011). Engineered Reversal of the beta oxidation cycle for the Synthesis of Fuels and Chemicals. Nature 476,355-359
- ↑ Patnaik, R. and Liao, J. (1994). "Engineering of Escherichia coli central metabolism for aromatic metabolite production with near theoretical yield". Appl. Environ. Microbiol. 60(11):3903-3908
- ↑ Keasling D.,Jay(2010). Advanced Biofuel production in microbes. Biotechnol.J.,5,147-162
- ↑ University of California - Los Angeles (2008, December 18). "Genetic Modification Turns E. Coli Bacteria Into High Density Biofuel". ScienceDaily. Retrieved December 7, 2011, from http://www.sciencedaily.com/releases/2008/12/081218151652.htm
- ↑ Schellenberger, J., Que, R., Fleming, R., et al. (2011). "Quantitative prediction of cellular metabolism with constraint-based models: the COBRA Toolbox v2.0". Nature Protocols. 6(9):1290-1307
- ↑ Rocha, I., Maia, P., Evangelista, P., et al. (2010). "OptFlux: an open-source software platform for in silico metabolic engineering". BMC Sys Biol. 45(4)
- ↑ Work, T.S., Hinton, R., Work, E., Dobrota, M., Chard, T. (1980). "Laboratory Techniques in Biochemistry and Molecular Biology". v.8
- ↑ Wiechert, W. and de Graaf, A.A. (2000). "Bidirectional Reaction Steps in Metabolic Networks: Modeling and Simulation of Carbon Isotope Labeling Experiments". Biotechnol. Bioeng. 55(1):101-117
External links
Biotechnology Industry Organization(BIO) website:
See also
- Bioreactor
- Bacterial transformation
- Genetic engineering