Hydrophobicity scales
Hydrophobicity scales are values that define relative hydrophobicity of amino acid residues. The more positive the value, the more hydrophobic are the amino acids located in that region of the protein. These scales are commonly used to predict the transmembrane alpha-helices of membrane proteins. When consecutively measuring amino acids of a protein, changes in value indicate attraction of specific protein regions towards the hydrophobic region inside lipid bilayer.
Hydrophobicity and the hydrophobic effect

The hydrophobic effect represents the tendency of water to exclude non-polar molecules. The effect originates from the disruption of highly dynamic hydrogen bonds between molecules of liquid water. Polar chemical groups, such as OH group in methanol do not cause the hydrophobic effect. However, a pure hydrocarbon molecule, for example hexane, cannot accept or donate hydrogen bonds to water. Introduction of hexane into water causes disruption of the hydrogen bonding network between water molecules. The hydrogen bonds are partially reconstructed by building a water "cage" around the hexane molecule, similar to that in clathrate hydrates formed at lower temperatures. The mobility of water molecules in the "cage" (or solvation shell) is strongly restricted. This leads to significant losses in translational and rotational entropy of water molecules and makes the process unfavorable in terms of free energy of the system.[1][1][2][3][4]
Types of amino acid hydrophobicity scales

A number of different hydrophobicity scales have been developed.[2][5][6][7]
There are clear differences between the four scales shown in the table.[8] Both the second and fourth scales place cysteine as the most hydrophobic residue, unlike the other two scales. This difference is due to the different methods used to measure hydrophobicity. The method used to obtain the Janin and Rose et al. scales was to examine proteins with known 3-D structures and define the hydrophobic character as the tendency for a residue to be found inside of a protein rather than on its surface.[9][10] Since cysteine forms disulfide bonds that must occur inside a globular structure, cysteine is ranked as the most hydrophobic. The first and third scales are derived from the physiochemical properties of the amino acid side chains. These scales result mainly from inspection of the amino acid structures.[11][12] Biswas et al., divided the scales based on the method used to obtain the scale into five different categories.[2]
Partitioning methods
The most common method of measuring amino acid hydrophobicity is partitioning between two immiscible liquid phases. Different organic solvents are most widely used to mimic the protein interior. However, organic solvents are slightly miscible with water and the characteristics of both phases change making it difficult to obtain pure hydrophobicity scale.[13] Nozaki and Tanford proposed the first major hydrophobicity scale for nine amino acids.[14] Ethanol and dioxane are used as the organic solvents and the free energy of transfer of each amino acid was calculated. Non liquid phases can also be used with partitioning methods such as micellar phases and vapor phases. Two scales have been developed using micellar phases.[15][16] Fendler et al. measured the partitioning of 14 radiolabeled amino acids using sodium dodecyl sulfate (SDS) micelles. Also, amino acid side chain affinity for water was measured using vapor phases.[17] Vapor phases represent the simplest non polar phases, because it has no interaction with the solute.[18] The hydration potential and its correlation to the appearance of amino acids on the surface of proteins was studied by Wolfenden. Aqueous and polymer phases were used in the development of a novel partitioning scale.[19] Partitioning methods have many drawbacks. First, it is difficult to mimic the protein interior.[20][21] In addition, the role of self solvation makes using free amino acids very difficult. Moreover, hydrogen bonds that are lost in the transfer to organic solvents are not reformed but often in the interior of protein.[22]
Accessible surface area methods
Hydrophobicity scales can also be obtained by calculating the solvent accessible surface areas for amino acid residues in the expended polypeptide chain[22] or in alpha-helix and multiplying the surface areas by the empirical solvation parameters for the corresponding types of atoms.[2] A differential solvent accessible surface area hydrophobicity scale based on proteins as compacted networks near a critical point, due to self-organization by evolution, was constructed based on asymptotic power-law (self-similar) behavior.[23] This scale is based on a bioinformatic survey of 5526 high-resolution structures from the Protein Data Bank. This differential scale has two comparative advantages: (1) it is especially useful for treating changes in water-protein interactions that are too small to be accessible to conventional force-field calculations, and (2) for homologous structures, it can yield correlations with changes in properties from mutations in the amino acid sequences alone,without determining corresponding structural changes, either in vitro or in vivo.
Chromatographic methods
Reversed phase liquid chromatography (RPLC) is the most important chromatographic method for measuring solute hydrophobicity.[2][24] The non polar stationary phase mimics biological membranes. Peptide usage has many advantages because partition is not extended by the terminal charges in RPLC. Also, secondary structures formation is avoided by suing short sequence peptides. Derivatization of amino acids is necessary to ease its partition into a C18 bonded phase. Another scale had been developed in 1971 and used peptide retention on hydrophillic gel.[25] 1-butanol and pyridine were used as the mobile phase in this particular scale and glycine was used as the reference value. Pliska and his cowrkers [26] used thin layer chromatography to relate mobility values of free amino acids to their hydrophobicities. About a decade ago, another hydrophilicity scale was published, this scale used normal phase liquid chromatography and showed the retention of 121 peptides on an amide-80 column.[27] The absolute values and relative rankings of hydrophobicity determined by chromatographic methods can be affected by a number of parameters. These parameters include the silica surface area and pore diameter, the choice and PH of aqueous buffer, temperature and the bonding density of stationary phase chains.[2]
Site-directed mutagenesis
This method use DNA recombinant technology and it gives an actual measurement of protein stability. In his detailed site directed mutagenesis studies, Utani and his coworkers substituted 19 amino acids at Trp49 of the tryptophan synthase and he measured the free energy of unfolding. Interestingly, they found that the increased stability is directly proportional to increase in hydrophobicity up to a certain size limit. The main disadvantage of site directed mutagenesis method is that not all the 20 naturally occurring amino acids can substitute a single residue in a protein. Moreover, these methods have cost problems and is useful only for measuring protein stability.[2][28]
Physical property methods
The hydrophobicity scales developed by physical property methods are based on the measurement of different physical properties. Examples include, partial molar heat capacity, transition temperature and surface tension. Physical methods are easy to use and flexible in terms of solute. The most popular hydrophobicity scale was developed by measuring surface tension values for the naturally occurring 20 amino acids in NaCl solution.[29] The main drawbacks of surface tension measurements is that the broken hydrogen bonds and the neutralized charged groups remain at the solution air interface.[2][30] Another physical property method involve measuring the solvation free energy.[31] The solvation free energy is estimated as a product of an accessibility of an atom to the solvent and an atomic solvation parameter. Results indicate the solvation free energy lowers by an average of 1 Kcal/residue upon folding.[2]
Recent applications
Palliser and Parry have examined about 100 scales and found that they can use them for locating B-strands on the surface of proteins.[32] Hydrophobicity scales were also used to predict the preservation of the genetic code.[33] Trinquier observed a new order of the bases that better reflect the conserved character of the genetic code.[2] They believed new ordering of the bases was uracil-guanine-cystosine-adenine(UGCA)better reflected the conserved character of the genetic code compared to the commonly seen ordering UCAG.[2]
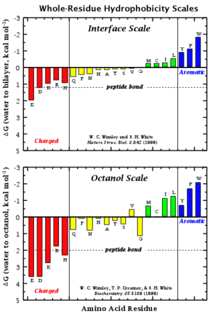
Wimley–White whole residue hydrophobicity scales
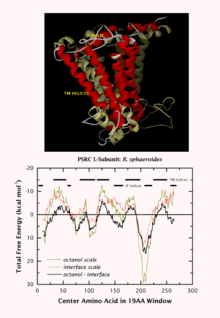
The Wimley–White whole residue hydrophobicity scales are significant for two reasons. First, they include the contributions of the peptide bonds as well as the sidechains, providing absolute values. Second, they are based on direct, experimentally determined values for transfer free energies of polypeptides. Two whole-residue hydrophobicity scales have been measured: One for the transfer of unfolded chains from water to the bilayer interface (referred to as the Wimley–White interfacial hydrophobicity scale) and one for the transfer of unfolded chains into octanol, which is relevant to the hydrocarbon core of a bilayer. The Stephen H. White website[34] provides an example of whole residue hydrophobicity scales showing the free energy of transfer ΔG(kcal/mol) from water to POPC interface and to n-octanol.[34] These two scales are then used together to make Whole residue hydropathy plots.[34] The hydropathy plot constructed using ΔGwoct − ΔGwif shows favorable peaks on the absolute scale that correspond to the known TM helices. Thus, the whole residue hydropathy plots proved the fact that transmembrane segments prefer a transmembrane location rather than a surface one.[35][36][37][38]
See also
References
- ↑ 1.0 1.1 Tanford,C., The hydrophobic effect(New York:Wiley.1980).
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 2.9 2.10 Kallol M. Biswas, Daniel R. DeVido, John G. Dorsey(2003) Journal of Chromatography A,1000, 637–655.
- ↑ W . Kauzmann, Adv. Protein Chem. 14 (1959) 1.
- ↑ Charton,M. and Charton,B.I. (1982) J. Theor. Biol., 99, 629–644.
- ↑ Kyte J, Doolittle RF (May 1982). "A simple method for displaying the hydropathic character of a protein". J. Mol. Biol. 157 (1): 105–32. doi:10.1016/0022-2836(82)90515-0. PMID 7108955.
- ↑ Eisenberg D (July 1984). "Three-dimensional structure of membrane and surface proteins". Ann. Rev. Biochem. 53: 595–623. doi:10.1146/annurev.bi.53.070184.003115. PMID 6383201.
- ↑ Rose,G.D. and Wolfenden,R. (1993) Annu. Rev. Biomol. Struct., 22, 381–415.
- ↑ http://prowl.rockefeller.edu/aainfo/hydro.htm
- ↑ J. Janin, Nature, 277(1979)491-492.
- ↑ G. Rose, A. Geselowitz, G. Lesser, R. Lee and M. Zehfus, Science 229(1985)834-838.
- ↑ R. Wolfenden, L. Andersson, P. Cullis and C. Southgate, Biochemistry 20(1981)849-855.
- ↑ J. Kyte and R. Doolittle, J. Mol Biol. 157(1982)105-132
- ↑ Kallol M. Biswas, Daniel R. DeVido, John G. Dorsey(2003) Journal of Chromatography A,1000, 637–655.
- ↑ Y . Nozaki, C. Tanford, J. Biol. Chem. 246 (1971) 2211.
- ↑ J .H. Fendler, F. Nome, J. Nagyvary, J. Mol. Evol. 6 (1975)215
- ↑ E .B. Leodidas, A.T. Hatton, J. Phys. Chem. 94 (1990) 6411.
- ↑ R . Wolfenden, L. Andersson, P.M. Cullis, C.C.B. Southgate, Biochemistry 20 (1981) 849. .
- ↑ K .A. Sharp, A. Nicholls, R. Friedman, B. Honig, Biochemistry 30 (1991) 9686.
- ↑ B .Y. Zaslavsky, N.M. Mestechkina, L.M. Miheeva, S.V. Rogozhin, J. Chromatogr. 240 (1982) 21.
- ↑ S . Damadoran, K.B. Song, J. Biol. Chem. 261 (1986) 7220.
- ↑ A . Ben-Naim, Biopolymers 29 (1990) 567.
- ↑ 22.0 22.1 C . Chothia, J. Mol. Biol. 105 (1976) 1.
- ↑ M. A. Moret, G. F. Zebende, Phys. Rev. E 75 (2007) 011920; J. C. Phillips, Phys. Rev. E 80 (2009) 051916.
- ↑ R .S. Hodges, B.-Y. Shu, N.E. Zhou, C.T. Mant, J. Chromatogr. A 676 (1994) 3.
- ↑ A .A. Aboderin, Int. J. Biochem. 2 (1971) 537.
- ↑ V . Pliska, M. Schmidt, J.L. Fauchere, J. Chromatogr. 216(1981) 79.
- ↑ M . Plass, K. Valko, M.H. Abraham, J. Chromatogr. A 803(1998) 51–60.
- ↑ K . Yutani, K. Ogasahara, T. Tsujita, Y. Sugino, Proc. Natl.Acad. Sci. USA 84 (1987) 4441.
- ↑ H .B. Bull, K. Breese, Arch. Biochem. Biophys. 161(1974)665
- ↑ J . Kyte, R.F. Doolittle, J. Mol. Biol. 157 (1982) 105.
- ↑ D . Eisenberg, A.D. McLachlan, Nature 319 (1986) 199
- ↑ C .C. Palliser, D.A.D. Parry, Proteins Struct. Funct. Genet. 42 (2001) 243.
- ↑ G . Trinquier, Y.-H. Sanejouand, Protein Eng. 11 (1998) 153.
- ↑ 34.0 34.1 34.2 White, Stephen (2006-06-29). "Experimentally Determined Hydrophobicity Scales". University of California, Irvine. Retrieved 2009-06-12.
- ↑ Wimley WC & White SH (1996). Nature Struct. Biol. 3:842-848.
- ↑ Wimley WC, Creamer TP & White SH (1996). Biochemistry 35:5109-5124.
- ↑ White SH. & Wimley WC (1998). Biochim. Biophys. Acta 1376:339-352.
- ↑ White SH & Wimley WC (1999). Annu. Rev. Biophys. Biomol. Struc. 28:319-365
External links
- ProtScale (web-based tool for calculating hydropathy plots)
- NetSurfP - Secondary Structure and Surface accessibility predictor
- Whole residue hydrophobicity scale
- Membrane protein explorer