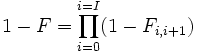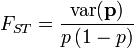F-statistics
In population genetics, F-statistics (also known as fixation indices) describe the statistically expected level of heterozygosity in a population; more specifically the expected degree of (usually) a reduction in heterozygosity when compared to Hardy–Weinberg expectation.
F-statistics can also be thought of as a measure of the correlation between genes drawn at different levels of a (hierarchically) subdivided population. This correlation is influenced by several evolutionary processes, such as mutation, migration, inbreeding, natural selection, or the Wahlund effect, but it was originally designed to measure the amount of allelic fixation owing to genetic drift.
The concept of F-statistics was developed during the 1920s by the American geneticist Sewall Wright,[1][2] who was interested in inbreeding in cattle. However, because complete dominance causes the phenotypes of homozygote dominants and heterozygotes to be the same, it was not until the advent of molecular genetics from the 1960s onwards that heterozygosity in populations could be measured.
F can be used to define effective population size.
Definitions and equations
The measures FIS, Fst, and FIT are related to the amounts of heterozygosity at various levels of population structure. Together, they are called F-statistics, and are derived from F, the inbreeding coefficient. In a simple two-allele system with inbreeding, the genotypic frequencies are:
The value for F is found by solving the equation for F using heterozygotes in the above inbred population. This becomes one minus the observed number of heterozygotes in a population divided by its expected number of heterozygotes at Hardy–Weinberg equilibrium:
where the expected value at Hardy–Weinberg equilibrium is given by
where p and q are the allele frequencies of A and a, respectively. It is also the probability that at any locus, two alleles from a random individuum of the population are identical by descent.
For example, consider the data from E.B. Ford (1971) on a single population of the scarlet tiger moth:
| Genotype | White-spotted (AA) | Intermediate (Aa) | Little spotting (aa) | Total |
|---|---|---|---|---|
| Number | 1469 | 138 | 5 | 1612 |
From this, the allele frequencies can be calculated, and the expectation of ƒ(AA) derived:
The different F-statistics look at different levels of population structure. FIT is the inbreeding coefficient of an individual (I) relative to the total (T) population, as above; FIS is the inbreeding coefficient of an individual (I) relative to the subpopulation (S), using the above for subpopulations and averaging them; and FST is the effect of subpopulations (S) compared to the total population (T), and is calculated by solving the equation:
as shown in the next section.
Partition due to population structure

Consider a population that has a population structure of two levels; one from the individual (I) to the subpopulation (S) and one from the subpopulation to the total (T). Then the total F, known here as FIT, can be partitioned into FIS (or f) and FST (or θ):
This may be further partitioned for population substructure, and it expands according to the rules of binomial expansion, so that for I partitions:
A reformulation of the definition of F would be the ratio of the average number of differences between pairs of chromosomes sampled within diploid individuals with the average number obtained when sampling chromosomes randomly from the population (excluding the grouping per individual). One can modify this definition and consider a grouping per sub-population instead of per individual. Population geneticists have used that idea to measure the degree of structure in a population.
Unfortunately, there is a large number of definitions for FST, causing some confusion in the scientific literature. A common definition is the following:
where the variance of p is computed across sub-populations and p(1 −p) is the expected frequency of heterozygotes.
 in human populations
in human populations
It is well established that the genetic diversity among human populations is low,[3] although the distribution of the genetic diversity was only roughly estimated. Early studies argued that 85-90% of the genetic variation is found within individuals residing in the same populations within continents (intra-continental populations) and only an additional 10-15% is found between populations of different continents (continental populations).[4][5][6][7][8] Later studies based on hundreds of thousands single-nucleotide polymorphism (SNPs) suggested that the genetic diversity between continental populations is even smaller and accounts for 3 to 7%[9][10][11][12][13][14] Most of these studies have used the FST statistics [15] or closely related statistics.[16][17]
See also
- Malecot's method of coancestry
- Heterozygosity
- Fixation index
References
- ↑ Wright, S (1950). "Genetical structure of populations". Nature 166 (4215): 247–9. Bibcode:1950Natur.166..247W. doi:10.1038/166247a0. PMID 15439261.
- ↑ Kulig, K (1985). "Utilization of emergency toxicology screens". The American journal of emergency medicine 3 (6): 573–4. LCCN 67025533. PMID 4063030.
- ↑ Holsinger, Kent E.; Weir, Bruce S. (2009). "Genetics in geographically structured populations: Defining, estimating and interpreting FST". Nature Reviews Genetics 10 (9): 639–50. doi:10.1038/nrg2611. PMID 19687804.
- ↑ Lewontin (1972). "The apportionment of human diversity". Evolutionary Biology 6: 381–98. doi:10.1007/978-1-4684-9063-3_14. ISBN 978-1-4684-9065-7.
- ↑ Bowcock, Anne M.; Kidd, Judith R.; Mountain, Joanna L.; Herbert, Joan M.; Carotenuto, Luciano; Kidd, Kenneth K.; Cavalli-Sforza, Luca (1991). "Drift, admixture, and selection in human evolution: A study with DNA polymorphisms". Proceedings of the National Academy of Sciences 88 (3): 839–43. Bibcode:1991PNAS...88..839B. doi:10.1073/pnas.88.3.839. JSTOR 2356081. PMC 50909. PMID 1992475.
- ↑ Barbujani, Guido; Magagni, Arianna; Minch, Eric; Cavalli-Sforza, L. Luca (1997). "An apportionment of human DNA diversity". Proceedings of the National Academy of Sciences of the United States of America 94 (9): 4516–9. Bibcode:1997PNAS...94.4516B. doi:10.1073/pnas.94.9.4516. JSTOR 42042. PMC 20754. PMID 9114021.
- ↑ Jorde, L.B.; Watkins, W.S.; Bamshad, M.J.; Dixon, M.E.; Ricker, C.E.; Seielstad, M.T.; Batzer, M.A. (2000). "The Distribution of Human Genetic Diversity: A Comparison of Mitochondrial, Autosomal, and Y-Chromosome Data". The American Journal of Human Genetics 66 (3): 979–88. doi:10.1086/302825. PMC 1288178. PMID 10712212.
- ↑ Jorde, Lynn B; Wooding, Stephen P (2004). "Genetic variation, classification and 'race'". Nature Genetics 36 (11s): S28–33. doi:10.1038/ng1435. PMID 15508000.
- ↑ Mahasirimongkol, Surakameth; Chantratita, Wasun; Promso, Somying; Pasomsab, Ekawat et al. (2006). "Similarity of the allele frequency and linkage disequilibrium pattern of single nucleotide polymorphisms in drug-related gene loci between Thai and northern East Asian populations: Implications for tagging SNP selection in Thais". Journal of Human Genetics 51 (10): 896–904. doi:10.1007/s10038-006-0041-1. PMID 16957813.
- ↑ Hannelius, Ulf; Salmela, Elina; Lappalainen, Tuuli; Guillot, Gilles; Lindgren, Cecilia M; Von Döbeln, Ulrika; Lahermo, Päivi; Kere, Juha (2008). "Population substructure in Finland and Sweden revealed by the use of spatial coordinates and a small number of unlinked autosomal SNPs". BMC Genetics 9: 54. doi:10.1186/1471-2156-9-54. PMC 2527025. PMID 18713460.
- ↑ Lao, Oscar; Lu, Timothy T.; Nothnagel, Michael; Junge, Olaf et al. (2008). "Correlation between Genetic and Geographic Structure in Europe". Current Biology 18 (16): 1241–8. doi:10.1016/j.cub.2008.07.049. PMID 18691889.
- ↑ Biswas, Shameek; Scheinfeldt, Laura B.; Akey, Joshua M. (2009). "Genome-wide Insights into the Patterns and Determinants of Fine-Scale Population Structure in Humans". The American Journal of Human Genetics 84 (5): 641. doi:10.1016/j.ajhg.2009.04.015.
- ↑ Nelis, Mari; Esko, Tõnu; Mägi, Reedik; Zimprich, Fritz et al. (2009). "Genetic Structure of Europeans: A View from the North–East". In Fleischer, Robert C. PLoS ONE 4 (5): e5472. Bibcode:2009PLoSO...4.5472N. doi:10.1371/journal.pone.0005472. PMC 2675054. PMID 19424496.
- ↑ Reich, David; Thangaraj, Kumarasamy; Patterson, Nick; Price, Alkes L. et al. (2009). "Reconstructing Indian population history". Nature 461 (7263): 489–94. Bibcode:2009Natur.461..489R. doi:10.1038/nature08365. PMC 2842210. PMID 19779445.
- ↑ Wright, Sewall (1965). "The Interpretation of Population Structure by F-Statistics with Special Regard to Systems of Mating". Evolution 19 (3): 395–420. doi:10.2307/2406450. JSTOR 2406450.
- ↑ Shalev, B. A.; Dvorin, A.; Herman, R.; Katz, Z.; Bornstein, S. (1991). "Long-term goose breeding for egg production and crammed liver weight". British Poultry Science 32 (4): 703–9. doi:10.1080/00071669108417396. PMID 1933444.
- ↑ Excoffier, L; Smouse, PE; Quattro, JM (1992). "Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data". Genetics 131 (2): 479–91. PMC 1205020. PMID 1644282.
External links
- Shane's Simple Guide to F-Statistics
- Analyzing the genetic structure of populations
- Wahlund effect, Wright's F-statistics
- Worked example of calculating F-statistics from genotypic data
- IAM based F-statistics
- F-statistics for Population Genetics Eco-Tool
- Population Structure (slides)
| |||||||||||||||||||||||