Escherichia coli contains a number of small RNAs located in intergenic regions of its genome. The presence of at least 55 of these has been verified experimentally.[1] 275 potential sRNA-encoding loci were identified computationally using the QRNA program. These loci will include false positives, so the number of sRNA genes in E. coli is likely to be less than 275.[2] A computational screen based on promoter sequences recognised by the sigma factor sigma 70 and on Rho-independent terminators predicted 24 putative sRNA genes, 14 of these were verified experimentally by northern blotting. The experimentally verified sRNAs included the well characterised sRNAs RprA and RyhB. Many of the sRNAs identified in this screen, including RprA, RyhB, SraB and SraL, are only expressed in the stationary phase of bacterial cell growth.[3] A screen for sRNA genes based on homology to Salmonella and Klebsiella identified 59 candidate sRNA genes. From this set of candidate genes, microarray analysis and northern blotting confirmed the existence of 17 previously undescribed sRNAs, many of which bind to the chaperone protein Hfq and regulate the translation of RpoS (Sigma 38).[4]
See also
Gallery of E.coli small RNA secondary structure images |
|---|
|
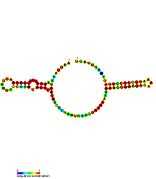
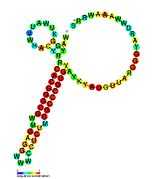
| sraA: Secondary structure taken from the Rfam database. Family RF02029 |
| tp2: Secondary structure taken from the Rfam database. Family RF02030 |
| tpke11: Secondary structure taken from the Rfam database. Family RF02031 |
|
|
|
References
- ↑ Hershberg R, Altuvia S, Margalit H (April 2003). "A survey of small RNA-encoding genes in Escherichia coli". Nucleic Acids Res. 31 (7): 1813–20. doi:10.1093/nar/gkg297. PMC 152812. PMID 12654996.
- ↑ Rivas E, Klein RJ, Jones TA, Eddy SR (September 2001). "Computational identification of noncoding RNAs in E. coli by comparative genomics". Curr. Biol. 11 (17): 1369–73. doi:10.1016/S0960-9822(01)00401-8. PMID 11553332.
- ↑ Argaman L, Hershberg R, Vogel J, et al. (June 2001). "Novel small RNA-encoding genes in the intergenic regions of Escherichia coli". Curr. Biol. 11 (12): 941–50. doi:10.1016/S0960-9822(01)00270-6. PMID 11448770.
- ↑ Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S (July 2001). "Identification of novel small RNAs using comparative genomics and microarrays". Genes Dev. 15 (13): 1637–51. doi:10.1101/gad.901001. PMC 312727. PMID 11445539.
Further reading
- Carter RJ, Dubchak I, Holbrook SR (October 2001). "A computational approach to identify genes for functional RNAs in genomic sequences". Nucleic Acids Res. 29 (19): 3928–38. doi:10.1093/nar/29.19.3928. PMC 60242. PMID 11574674.
- Chen S, Lesnik EA, Hall TA, et al. (2002). "A bioinformatics based approach to discover small RNA genes in the Escherichia coli genome". BioSystems 65 (2-3): 157–77. doi:10.1016/S0303-2647(02)00013-8. PMID 12069726.
- Tjaden B, Saxena RM, Stolyar S, Haynor DR, Kolker E, Rosenow C (September 2002). "Transcriptome analysis of Escherichia coli using high-density oligonucleotide probe arrays". Nucleic Acids Res. 30 (17): 3732–8. doi:10.1093/nar/gkf505. PMC 137427. PMID 12202758.
- Vogel J, Sharma CM (December 2005). "How to find small non-coding RNAs in bacteria". Biol. Chem. 386 (12): 1219–38. doi:10.1515/BC.2005.140. PMID 16336117.
- Raghavan, R.; Groisman, E. A.; Ochman, H. (2011). "Genome-wide detection of novel regulatory RNAs in E. Coli". Genome Research 21 (9): 1487–1497. doi:10.1101/gr.119370.110. PMC 3166833. PMID 21665928.