CpG island
| CpG island | |
|---|---|
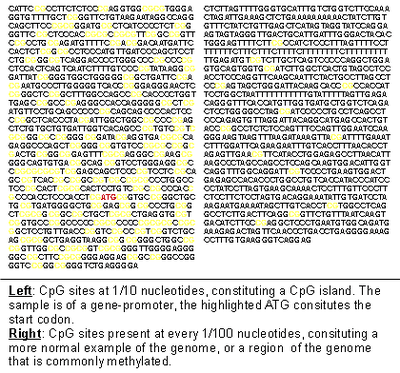 | |
 | |
| How methylation of CpG sites followed by spontaneous deamination leads to a lack of CpG sites in methylated DNA. As a result residual CpG islands are created in areas where methylation is rare, and CpG sites stick (or where C to T mutation is highly detrimental). | |
In genetics, CpG islands or CG islands (CGI) are genomic regions that contain a high frequency of CpG sites. The "p" in CpG refers to the phosphodiester bond between the cytosine and the guanine, which indicates that the C and the G are next to each other in sequence, regardless of being single- or double- stranded. In a CpG site, both C and G are found on the same strand of DNA or RNA and are connected by a phosphodiester bond. This is a covalent bond between atoms, stable and permanent as opposed to the three hydrogen bonds established after base-pairing of C and G in opposite strands of DNA.
However, objective definitions for CpG islands are limited. The usual formal definition of a CpG island is a region with at least 200 bp, and a GC percentage that is greater than 50%, and with an observed-to-expected CpG ratio that is greater than 60%. The "observed-to-expected CpG ratio" is calculated by formula ((Num of CpG/(Num of C × Num of G)) × Total number of nucleotides in the sequence).[1]

In mammalian genomes, CpG islands are typically 300-3,000 base pairs in length, and have been found in or near approximately 40% of promoters of mammalian genes.[2] About 70% of human promoters have a high CpG content. Given the frequency of GC two-nucleotide sequences, the number of CpG dinucleotides is much lower than would be expected.[3]
A 2002 study revised the rules of CpG island prediction to exclude other GC-rich genomic sequences such as Alu repeats. Based on an extensive search on the complete sequences of human chromosomes 21 and 22, DNA regions greater than 500 bp were found more likely to be the "true" CpG islands associated with the 5' regions of genes if they had a GC content greater than 55%, and an observed-to-expected CpG ratio of 65%.[4]
CpG islands are characterized by CpG dinucleotide content of at least 60% of that which would be statistically expected (~4–6%), whereas the rest of the genome has much lower CpG frequency (~1%), a phenomenon called CG suppression. Unlike CpG sites in the coding region of a gene, in most instances the CpG sites in the CpG islands of promoters are unmethylated if the genes are expressed. This observation led to the speculation that methylation of CpG sites in the promoter of a gene may inhibit gene expression. Methylation is central to imprinting, along with histone modifications.[5] Most of the methylation occurs a short distance from the CpG islands (at "CpG island shores") rather than in the islands themselves.[6]
CpG islands typically occur at or near the transcription start site of genes, particularly housekeeping genes, in vertebrates.[3] Normally a C (cytosine) base followed immediately by a G (guanine) base (a CpG) is rare in vertebrate DNA because the cytosines in such an arrangement tend to be methylated. This methylation helps distinguish the newly synthesized DNA strand from the parent strand, which aids in the final stages of DNA proofreading after duplication. However, over evolutionary time methylated cytosines tend to turn into thymines because of spontaneous deamination. While there is a special enzyme in human (Thymine-DNA glycosylase, or TDG) that specifically replaces T's from T/G mismatches, it is not sufficiently effective to prevent the relatively rapid mutation of the dinucleotides. The result is that CpGs are relatively rare. The existence of CpG islands is usually explained by the existence of selective forces for relatively high CpG content, or low levels of methylation in that genomic area, perhaps having to do with the regulation of gene expression. Recently a study showed that most CpG islands are a result of non-selective forces.[7]
Additional images
See also
References
- ↑ Gardiner-Garden M, Frommer M (1987). "CpG islands in vertebrate genomes". Journal of Molecular Biology 196 (2): 261–82. doi:10.1016/0022-2836(87)90689-9. PMID 3656447.
- ↑ Fatemi M, Pao MM, Jeong S, Gal-Yam EN, Egger G, Weisenberger DJ, Jones PA (2005). "Footprinting of mammalian promoters: use of a CpG DNA methyltransferase revealing nucleosome positions at a single molecule level". Nucleic Acids Res 33 (20): e176. doi:10.1093/nar/gni180. PMC 1292996. PMID 16314307.
- ↑ 3.0 3.1 Saxonov S, Berg P, Brutlag DL (2006). "A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters". Proc Natl Acad Sci USA 103 (5): 1412–1417. doi:10.1073/pnas.0510310103. PMC 1345710. PMID 16432200.
- ↑ Takai D, Jones PA (2002). "Comprehensive analysis of CpG islands in human chromosomes 21 and 22.". Proc Natl Acad Sci USA 99 (6): 3740–5. doi:10.1073/pnas.052410099. PMC 122594. PMID 11891299.
- ↑ Feil R, Berger F (2007). "Convergent evolution of genomic imprinting in plants and mammals". Trends Genet 23 (4): 192–199. doi:10.1016/j.tig.2007.02.004. PMID 17316885.
- ↑ Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, Ji H, Potash JB, Sabunciyan S, Feinberg AP (2009). "The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores". Nature Genetics 41 (2): 178–186. doi:10.1038/ng.298. PMC 2729128. PMID 19151715.
- ↑ Cohen N, Kenigsberg E, Tanay A (2011). "Primate CpG Islands Are Maintained by Heterogeneous Evolutionary Regimes Involving Minimal Selection". Cell 145 (5): 773–786. doi:10.1016/j.cell.2011.04.024. PMID 21620139.
External links
- M. D. Anderson Cancer Center, mdanderson.org
- CpG Island Searcher using the updated rules from Takai and Jones, cpgislands.usc.edu
- EMBOSS CpGPlot/CpGReport/Isochore, ebi.ac.uk
- Google Scholar Search for 'CpG Island', scholar.google.com.hk
- CpG Islands at the US National Library of Medicine Medical Subject Headings (MeSH)

