BLOSUM
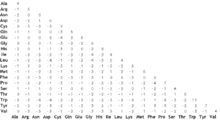
In bioinformatics, the BLOSUM (BLOcks SUbstitution Matrix) matrix is a substitution matrix used for sequence alignment of proteins. BLOSUM matrices are used to score alignments between evolutionarily divergent protein sequences. They are based on local alignments. BLOSUM matrices were first introduced in a paper by Henikoff and Henikoff.[1] They scanned the BLOCKS database for very conserved regions of protein families (that do not have gaps in the sequence alignment) and then counted the relative frequencies of amino acids and their substitution probabilities. Then, they calculated a log-odds score for each of the 210 possible substitution pairs of the 20 standard amino acids. All BLOSUM matrices are based on observed alignments; they are not extrapolated from comparisons of closely related proteins like the PAM Matrices.
Several sets of BLOSUM matrices exist using different alignment databases, named with numbers. BLOSUM matrices with high numbers are designed for comparing closely related sequences, while those with low numbers are designed for comparing distant related sequences. For example, BLOSUM80 is used for less divergent alignments, and BLOSUM45 is used for more divergent alignments. The matrices were created by merging (clustering) all sequences that were more similar than a given percentage into one single sequence and then comparing those sequences (that were all more divergent than the given percentage value) only; thus reducing the contribution of closely related sequences. The percentage used was appended to the name, giving BLOSUM80 for example where sequences that were more than 80% identical were clustered.
Scores within a BLOSUM are log-odds scores that measure, in an alignment, the logarithm for the ratio of the likelihood of two amino acids appearing with a biological sense and the likelihood of the same amino acids appearing by chance.[2] The matrices are based on the minimum percentage identity of the aligned protein sequence used in calculating them.[2] Every possible identity or substitution is assigned a score based on its observed frequences in the alignment of related proteins.[3] A positive score is given to the more likely substitutions while a negative score is given to the less likely substitutions.
To calculate a BLOSUM matrix, the following equation is used:
Here,  is the probability of two amino acids
is the probability of two amino acids  and
and  replacing each other in a homologous sequence, and
replacing each other in a homologous sequence, and  and
and 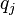 are the background probabilities of finding the amino acids
are the background probabilities of finding the amino acids  and
and  in any protein sequence. The factor
in any protein sequence. The factor 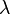 is a scaling factor, set such that the matrix contains easily computable integer values.
is a scaling factor, set such that the matrix contains easily computable integer values.
An article in Nature Biotechnology[4] revealed that the BLOSUM62 used for so many years as a standard is not exactly accurate according to the algorithm described by Henikoff and Henikoff.[1] Surprisingly, the miscalculated BLOSUM62 improves search performance.[4]
See also
References
- ↑ 1.0 1.1 Henikoff, S.; Henikoff, J.G. (1992). "Amino Acid Substitution Matrices from Protein Blocks". PNAS 89 (22): 10915–10919. doi:10.1073/pnas.89.22.10915. PMC 50453. PMID 1438297.
- ↑ 2.0 2.1 Albert Y. Zomaya (2006). Handbook of Nature-Inspired And Innovative Computing. New York, NY: Springer. ISBN 0-387-40532-1.page 673
- ↑ NIH "Scoring Systems"
- ↑ 4.0 4.1 Mark P Styczynski; Kyle L Jensen, Isidore Rigoutsos, Gregory Stephanopoulos (2008). "BLOSUM62 miscalculations improve search performance". Nat. Biotech. 26 (3): 274–275. doi:10.1038/nbt0308-274. PMID 18327232.
External links
- Sean R. Eddy (2004). "Where did the BLOSUM62 alignment score matrix come from?". Nature Biotechnology 22 (8): 1035. doi:10.1038/nbt0804-1035. PMID 15286655.
- BLOCKS WWW server
- Scoring systems for BLAST at NCBI
- Data files of BLOSUM on the NCBI FTP server.
- Interactive BLOSUM Network Visualization
