25-Hydroxyvitamin D3 1-alpha-hydroxylase
From Wikipedia, the free encyclopedia
25-Hydroxyvitamin D3 1-alpha-hydroxylase (VD3 1A hydroxylase) also known as cytochrome p450 27B1 (CYP27B1) is a cytochrome P450 enzyme that in humans is encoded by the CYP27B1 gene.[1][2][3]
VD3 1A hydroxylase is located in the proximal tubule of the kidney and a variety of other tissues, including skin (keratinocytes), immune cells,[4] and bone (osteoblasts).[5] The enzyme catalyzes the hydroxylation of Calcifediol to calcitriol (the bioactive form of Vitamin D):[6]
- calcidiol + NADPH + H+ + O2
 calcitriol + NADP+ + H2O
calcitriol + NADP+ + H2O
| ||||||||||||||||||||||||||||||||||||
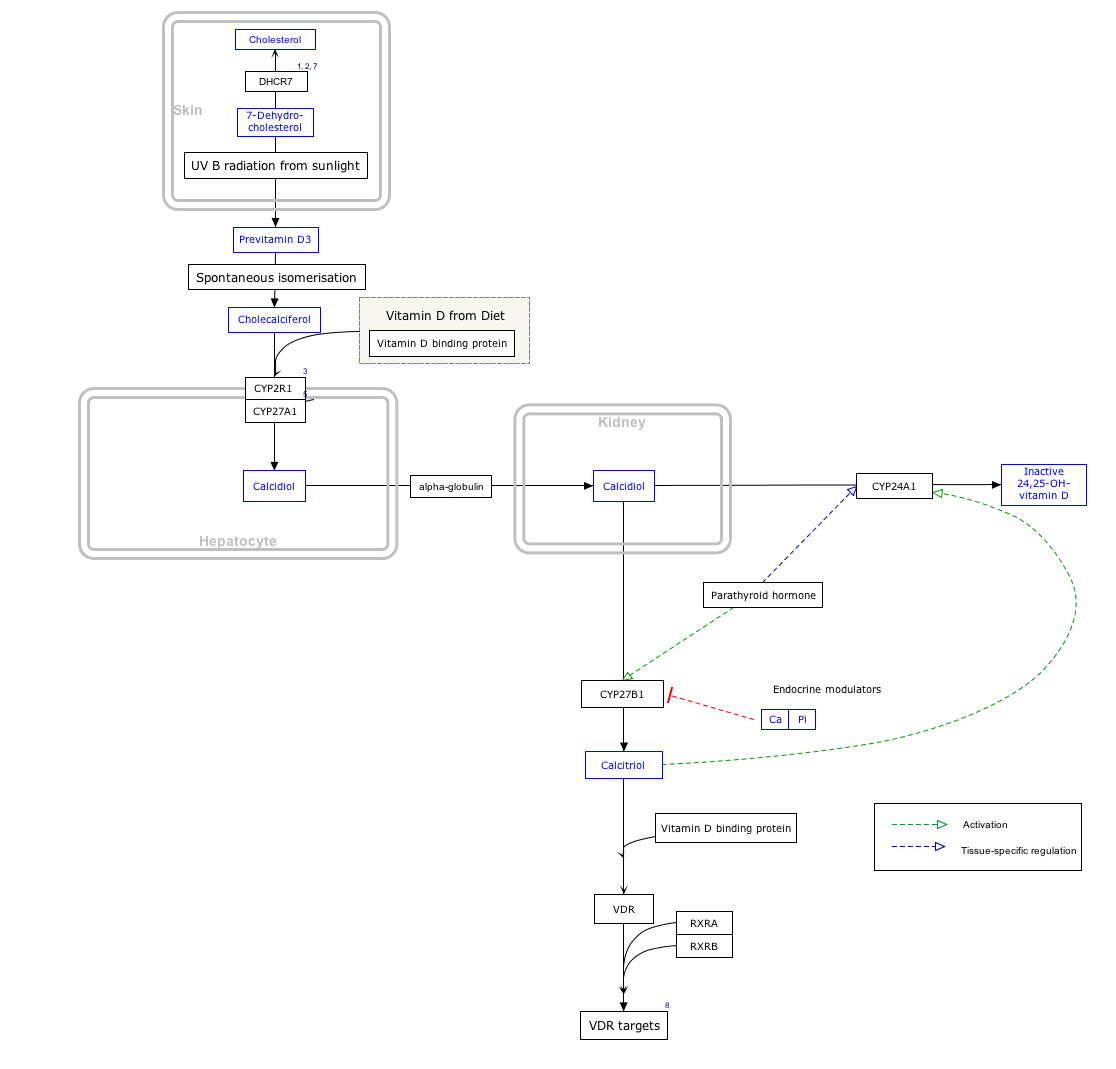
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles. [§ 1]
Vitamin D Synthesis Pathway edit
- ↑ The interactive pathway map can be edited at WikiPathways: "VitaminDSynthesis_WP1531".
References
- ↑ "Entrez Gene: cytochrome P450".
- ↑ Takeyama K, Kitanaka S, Sato T, Kobori M, Yanagisawa J, Kato S (September 1997). "25-Hydroxyvitamin D3 1alpha-hydroxylase and vitamin D synthesis". Science 277 (5333): 1827–30. doi:10.1126/science.277.5333.1827. PMID 9295274.
- ↑ Monkawa T, Yoshida T, Wakino S, Shinki T, Anazawa H, Deluca HF, Suda T, Hayashi M, Saruta T (October 1997). "Molecular cloning of cDNA and genomic DNA for human 25-hydroxyvitamin D3 1 alpha-hydroxylase". Biochem. Biophys. Res. Commun. 239 (2): 527–33. doi:10.1006/bbrc.1997.7508. PMID 9344864.
- ↑ Sigmundsdottir H, Pan J, Debes GF, Alt C, Habtezion A, Soler D, Butcher EC (March 2007). "DCs metabolize sunlight-induced vitamin D3 to 'program' T cell attraction to the epidermal chemokine CCL27". Nat. Immunol. 8 (3): 285–93. doi:10.1038/ni1433. PMID 17259988.
- ↑ Kogawa M, Findlay DM, Anderson PH, Ormsby R, Vincent C, Morris HA, Atkins GJ (October 2010). "Osteoclastic metabolism of 25(OH)-vitamin D3: a potential mechanism for optimization of bone resorption". Endocrinology 151 (10): 4613–25. doi:10.1210/en.2010-0334. PMID 20739402.
- ↑ Gray RW, Omdahl JL, Ghazarian JG, DeLuca HF (December 1972). "25-Hydroxycholecalciferol-1-hydroxylase. Subcellular location and properties". J. Biol. Chem. 247 (23): 7528–32. PMID 4404596.
Further reading
- Carr EJ, Niederer HA, Williams J et al. (2009). "Confirmation of the genetic association of CTLA4 and PTPN22 with ANCA-associated vasculitis". BMC Med. Genet. 10: 121. doi:10.1186/1471-2350-10-121. PMID 19951419.
- Alzahrani AS, Zou M, Baitei EY et al. (2010). "A novel G102E mutation of CYP27B1 in a large family with vitamin D-dependent rickets type 1". J. Clin. Endocrinol. Metab. 95 (9): 4176–83. doi:10.1210/jc.2009-2278. PMID 20534770.
- Lagishetty V, Chun RF, Liu NQ et al. (2010). "1alpha-hydroxylase and innate immune responses to 25-hydroxyvitamin D in colonic cell lines". J. Steroid Biochem. Mol. Biol. 121 (1–2): 228–33. doi:10.1016/j.jsbmb.2010.02.004. PMC 2891066. PMID 20152900.
- Giroux S, Elfassihi L, Clément V et al. (2010). "High-density polymorphisms analysis of 23 candidate genes for association with bone mineral density". Bone 47 (5): 975–81. doi:10.1016/j.bone.2010.06.030. PMID 20654748.
- Zhou S, LeBoff MS, Glowacki J (2010). "Vitamin D metabolism and action in human bone marrow stromal cells". Endocrinology 151 (1): 14–22. doi:10.1210/en.2009-0969. PMC 2803155. PMID 19966181.
- Payne AH, Hales DB (2004). "Overview of steroidogenic enzymes in the pathway from cholesterol to active steroid hormones". Endocr. Rev. 25 (6): 947–70. doi:10.1210/er.2003-0030. PMID 15583024.
- Maver A, Medica I, Salobir B et al. (2010). "Lack of association of immune-response-gene polymorphisms with susceptibility to sarcoidosis in Slovenian patients". Genet. Mol. Res. 9 (1): 58–68. doi:10.4238/vol9-1gmr682. PMID 20082271.
- Shen H, Bielak LF, Ferguson JF et al. (2010). "Association of the vitamin D metabolism gene CYP24A1 with coronary artery calcification". Arterioscler. Thromb. Vasc. Biol. 30 (12): 2648–54. doi:10.1161/ATVBAHA.110.211805. PMC 2988112. PMID 20847308.
- Sundqvist E, Bäärnhielm M, Alfredsson L et al. (2010). "Confirmation of association between multiple sclerosis and CYP27B1". Eur. J. Hum. Genet. 18 (12): 1349–52. doi:10.1038/ejhg.2010.113. PMC 3002863. PMID 20648053.
- Fichna M, Zurawek M, Januszkiewicz-Lewandowska D et al. (2010). "Association of the CYP27B1 C(-1260)A polymorphism with autoimmune Addison's disease". Exp. Clin. Endocrinol. Diabetes 118 (8): 544–9. doi:10.1055/s-0029-1241206. PMID 19998245.
- Holt SK, Kwon EM, Koopmeiners JS et al. (2010). "Vitamin D pathway gene variants and prostate cancer prognosis". Prostate 70 (13): 1448–60. doi:10.1002/pros.21180. PMC 2927712. PMID 20687218.
- Bu FX, Armas L, Lappe J et al. (2010). "Comprehensive association analysis of nine candidate genes with serum 25-hydroxy vitamin D levels among healthy Caucasian subjects". Hum. Genet. 128 (5): 549–56. doi:10.1007/s00439-010-0881-9. PMID 20809279.
- Fichna M, Zurawek M, Januszkiewicz-Lewandowska D et al. (2010). "PTPN22, PDCD1 and CYP27B1 polymorphisms and susceptibility to type 1 diabetes in Polish patients". Int. J. Immunogenet. 37 (5): 367–72. doi:10.1111/j.1744-313X.2010.00935.x. PMID 20518841.
- Wjst M, Heimbeck I, Kutschke D, Pukelsheim K (2010). "Epigenetic regulation of vitamin D converting enzymes". J. Steroid Biochem. Mol. Biol. 121 (1–2): 80–3. doi:10.1016/j.jsbmb.2010.03.056. PMID 20304056.
- Liu CY, Wu MC, Chen F et al. (2010). "A Large-scale genetic association study of esophageal adenocarcinoma risk". Carcinogenesis 31 (7): 1259–63. doi:10.1093/carcin/bgq092. PMC 2893800. PMID 20453000.
- Sunyer J, Basagaña X, González JR et al. (2010). "Early life environment, neurodevelopment and the interrelation with atopy". Environ. Res. 110 (7): 733–8. doi:10.1016/j.envres.2010.07.005. PMID 20701904.
- Simon KC, Munger KL, Xing Yang , Ascherio A (2010). "Polymorphisms in vitamin D metabolism related genes and risk of multiple sclerosis". Mult. Scler. 16 (2): 133–8. doi:10.1177/1352458509355069. PMC 2819633. PMID 20007432.
- Hendrickson SL, Lautenberger JA, Chinn LW et al. (2010). "Genetic variants in nuclear-encoded mitochondrial genes influence AIDS progression". In Badger, Jonathan H. PLoS ONE 5 (9): e12862. doi:10.1371/journal.pone.0012862. PMC 2943476. PMID 20877624.
- Dusso AS, Brown AJ, Slatopolsky E (2005). "Vitamin D". Am. J. Physiol. Renal Physiol. 289 (1): F8–28. doi:10.1152/ajprenal.00336.2004. PMID 15951480.
- Bailey SD, Xie C, Do R et al. (2010). "Variation at the NFATC2 locus increases the risk of thiazolidinedione-induced edema in the Diabetes REduction Assessment with ramipril and rosiglitazone Medication (DREAM) study". Diabetes Care 33 (10): 2250–3. doi:10.2337/dc10-0452. PMC 2945168. PMID 20628086.
External links
- 25-Hydroxyvitamin D3 1-alpha-Hydroxylase at the US National Library of Medicine Medical Subject Headings (MeSH)
| ||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
This article is issued from Wikipedia. The text is available under the Creative Commons Attribution/Share Alike; additional terms may apply for the media files.