Conformational isomerism
In chemistry, conformational isomerism is a form of stereoisomerism in which the isomers can be interconverted exclusively by rotations about formally single bonds.[1] Such isomers are generally referred to as conformational isomers or conformers and specifically as rotamers[2] when the rotation leading to different conformations is restricted (hindered) rotation,in the sense that there exists a rotational energy barrier that needs to be overcome to convert one conformer to another. Conformational isomers are thus distinct from the other classes of stereoisomers for which interconversion necessarily involves breaking and reforming of chemical bonds. The rotational barrier, or barrier to rotation, is the activation energy required to interconvert rotamers.
Contents |
Types of conformational isomerism
Butane has three rotamers: two gauche conformers, which are enantiomeric and an anti conformer, where the four carbon centres are coplanar. The three eclipsed conformations with dihedral angles of 0°,120° and 240° are not considered to be rotamers, but are instead transition states.
Some important examples of conformational isomerism include:
- Linear alkane conformations with staggered, eclipsed and gauche conformers, and
- Ring conformation
- Cyclohexane conformations with chair and boat conformers.
- Carbohydrate conformation
- Atropisomerism- due to restricted rotation about a bond, a molecule can become chiral
- Folding of molecules, where some shapes are stable and functional, but others are not.
Equilibrium population of conformers
The population of different conformers follows a Boltzmann distribution:
The left hand side is the equilibrium ratio of conformer i to the total. 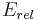 is the relative energy of the i-th conformer from the minimum energy conformer.
is the relative energy of the i-th conformer from the minimum energy conformer. 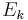 is the relative energy of the k-th conformer from the minimum energy conformer. R is the molar ideal gas constant equal to 8.31 J/(mol·K) and T is the temperature in kelvins (K). The denominator of the right side is the partition function.
is the relative energy of the k-th conformer from the minimum energy conformer. R is the molar ideal gas constant equal to 8.31 J/(mol·K) and T is the temperature in kelvins (K). The denominator of the right side is the partition function.
Isolation or observation of the conformational isomers
- Atropoisomers can be quite stable depending on the steric effects and were the first conformational isomers to be identified.[3] In the biphenylic system atropisomerism is especially prevalent, e.g. binaphthol.
- In cyclohexane derivatives, the two chair conformers interconvert with rates on the order of 105ring flips/sec, which obviously precludes their separation.[3] The equatorial conformer crystallizes selectively, and when these crystals are dissolved at very low temperatures, one can directly monitor the approach to equilibrium by NMR spectroscopy.[4]
Techniques for study of conformational isomerism
Most information on conformational isomerism comes from single crystal X-ray diffraction studies. IR spectroscopy is ordinarily used to measure conformer ratios. For the axial and equatorial conformer of bromocyclohexane, νCBr differs by almost 50 cm−1.[3]
The dynamics of conformational (and other kinds of) isomerism can be monitored by NMR spectroscopy at varying temperatures. The technique applies to barriers of 8-14 kcal/mol, and species exhibiting such dynamics are often called "fluxional".
Conformation-dependent reactions
Reaction rates are highly dependent on the conformation of the reactants. This theme is especially well elucidated in organic chemistry. One example is provided by the elimination reactions, which involve the simultaneous removal of a proton and a leaving group from vicinal positions under the influence of a base. The mechanism requires that the departing atoms or groups follow antiparallel trajectories. For open chain substrates this geometric prerequisite is met by at least one of the three staggered conformers. For some cyclic substrates, however, an antiparallel arrangement may not be attainable depending. Adjacent substituents on a cyclohexane ring can achieve antiperiplanarity only when they occupy trans diaxial positions. One consequence of this analysis is that trans-4-tert-butylcyclohexyl chloride cannot easily eliminate but instead undergoes substitution.
Protein rotamer libraries
A rotamer library is a collection of rotamers for each residue type in proteins with side-chain degrees of freedom. Rotamer libraries usually contain information about both conformation and frequency of a certain conformation. Often libraries will also contain information about the variance about dihedral angle means or modes, which can be used in sampling.[4]
Side-chain dihedral angles are not evenly distributed, but for most side chain types, the 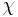 angles occur in tight clusters around certain values. Rotamer libraries therefore are usually derived from statistical analysis of side-chain conformations in known structures of proteins by clustering observed conformations or by dividing dihedral angle space into bins, and determining an average conformation in each bin. This division is usually on physical-chemical grounds, as in the divisions for rotation about sp3-sp3 bonds into three 120° bins centered on each staggered conformation (60°, 180°, -60°).
angles occur in tight clusters around certain values. Rotamer libraries therefore are usually derived from statistical analysis of side-chain conformations in known structures of proteins by clustering observed conformations or by dividing dihedral angle space into bins, and determining an average conformation in each bin. This division is usually on physical-chemical grounds, as in the divisions for rotation about sp3-sp3 bonds into three 120° bins centered on each staggered conformation (60°, 180°, -60°).
Rotamer libraries can be backbone-independent, secondary-structure-dependent, or backbone-dependent. The distinctions are made depending on whether the dihedral angles for the rotamers and/or their frequencies depend on the local backbone conformation or not. Backbone-independent rotamer libraries make no reference to backbone conformation, and are calculated from all available side chains of a certain type.[5] Secondary-structure-dependent libraries present different dihedral angles and/or rotamer frequencies for 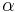 -helix,
-helix, 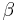 -sheet, or coil secondary structures. Backbone-dependent rotamer libraries present conformations and/or frequencies dependent on the local backbone conformation as defined by the backbone dihedral angles
-sheet, or coil secondary structures. Backbone-dependent rotamer libraries present conformations and/or frequencies dependent on the local backbone conformation as defined by the backbone dihedral angles 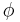 and
and 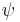 , regardless of secondary structure.[6] Finally, a variant on backbone-dependent rotamer libraries exists in the form of position-specific rotamers, those defined by a fragment usually of 5 amino acids in length, where the central residue’s side chain conformation is examined.
, regardless of secondary structure.[6] Finally, a variant on backbone-dependent rotamer libraries exists in the form of position-specific rotamers, those defined by a fragment usually of 5 amino acids in length, where the central residue’s side chain conformation is examined.
See also
References
- ^ IUPAC definition of a conformer.
- ^ IUPAC, Compendium of Chemical Terminology, 2nd ed. (the "Gold Book") (1997). Online corrected version: (1996) "Rotamer".
- ^ a b c Eliel, E. L.; Wilen, S. H.; Mander, L. N. "Stereochemistry Of Organic Compounds", J. Wiley and Sons, 1994. ISBN 0-471-01670-5.
- ^ a b Dunbrack, R. (2002). "Rotamer Libraries in the 21st Century". Current Opinion in Structural Biology 12 (4): 431–440. doi:10.1016/S0959-440X(02)00344-5. PMID 12163064.
- ^ Richardson Rotamer Libraries
- ^ Dunbrack Rotamer Libraries