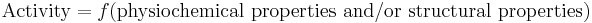Quantitative structure–activity relationship
Quantitative structure–activity relationship (QSAR) or QSPR (quantitative structure–property relationship) is the process by which chemical structure is quantitatively correlated with a well defined process, such as biological activity or chemical reactivity.
For example, biological activity can be expressed quantitatively as the concentration of a substance required to give a certain biological response. Additionally, when physicochemical properties or structures are expressed by numbers, one can form a mathematical relationship, or quantitative structure-activity relationship, between the two. The mathematical expression can then be used to predict the biological response of other chemical structures.
QSAR's most general mathematical form is:
Contents |
SAR and the SAR paradox
The basic assumption for all molecule based hypotheses is that similar molecules have similar activities. This principle is also called Structure–Activity Relationship (SAR). The underlying problem is therefore how to define a small difference on a molecular level, since each kind of activity, e.g. reaction ability, biotransformation ability, solubility, target activity, and so on, might depend on another difference. A good example was given in the bioisosterism review of Patanie/LaVoie.[1]
In general, one is more interested in finding strong trends. Created hypotheses usually rely on a finite number of chemical data. Thus, the induction principle should be respected to avoid overfitted hypotheses and deriving overfitted and useless interpretations on structural/molecular data.
The SAR paradox refers to the fact that it is not the case that all similar molecules have similar activities.
Types
Fragment based (group contribution)
It has been shown that the logP of compound can be determined by the sum of its fragments. Fragmentary logP values have been determined statistically. This method gives mixed results and is generally not trusted to have accuracy of more than ±0.1 units.[2]
Group or Fragment based QSAR is also known as GQSAR. GQSAR allows flexibility to study various molecular fragments of interest in relation to the variation in biological response. The molecular fragments could be substituents at various substitution sites in congeneric set of molecules or could be on the basis of pre-defined chemical rules in case of non-congeneric set. GQSAR also considers cross-terms fragment descriptors, which could be helpful in identification of key fragment interactions in determining variation of activity.[3] Lead discovery using Fragnomics is an emerging paradigm. In this context FB-QSAR proves to be a promising strategy for fragment library design and in fragment-to-lead identification endeavours.[4]
3D-QSAR
3D-QSAR refers to the application of force field calculations requiring three-dimensional structures, e.g. based on protein crystallography or molecule superimposition. It uses computed potentials, e.g. the Lennard-Jones potential, rather than experimental constants and is concerned with the overall molecule rather than a single substituent. It examines the steric fields (shape of the molecule) and the electrostatic fields based on the applied energy function.[5]
The created data space is then usually reduced by a following feature extraction (see also dimensionality reduction). The following learning method can be any of the already mentioned machine learning methods, e.g. support vector machines.[6] An alternative approach uses multiple-instance learning by encoding molecules as sets of data instances, each of which represents a possible molecular conformation. A label or response is assigned to each set corresponding to the activity of the molecule, which is assumed to be determined by at least one instance in the set (i.e. some conformation of the molecule).[7]
In the literature it can be often found that chemists have a preference for partial least squares (PLS) methods, since it applies the feature extraction and induction in one step.
On June 18th 2011 the CoMFA patent has dropped any restriction on the use of GRID and PLS technologies and the RCMD team (www.rcmd.it) has opened a 3D QSAR web server (www.3d-qsar.com).
Data mining
For the coding usually a relatively large number of features or molecular descriptors are calculated, which can lack structural interpretation ability. In combination with the later applied learning method or as preprocessing step occurs a feature selection problem.
A typical data mining based prediction uses e.g. support vector machines, decision trees, neural networks for inducing a predictive learning model.
Molecule mining approaches, a special case of structured data mining approaches, apply a similarity matrix based prediction or an automatic fragmentation scheme into molecular substructures. Furthermore there exist also approaches using maximum common subgraph searches or graph kernels.[8][9]
Judging the quality of QSAR models
QSARs represent predictive models derived from application of statistical tools correlating biological activity (including desirable therapeutic effect and undesirable side effects) of chemicals (drugs/toxicants/environmental pollutants) with descriptors representative of molecular structure and/or properties. QSARs are being applied in many disciplines for example risk assessment, toxicity prediction, and regulatory decisions[10] in addition to drug discovery and lead optimization.[11] Obtaining a good quality QSAR model depends on many factors, such as the quality of biological data, the choice of descriptors and statistical methods. Any QSAR modeling should ultimately lead to statistically robust models capable of making accurate and reliable predictions of biological activities of new compounds.
For validation of QSAR models usually four strategies are adopted:[12]
- internal validation or cross-validation;
- validation by dividing the data set into training and test compounds;
- true external validation by application of model on external data and
- data randomization or Y-scrambling.
The success of any QSAR model depends on accuracy of the input data, selection of appropriate descriptors and statistical tools, and most importantly validation of the developed model. Validation is the process by which the reliability and relevance of a procedure are established for a specific purpose.[13] Leave one-out cross-validation generally leads to an overestimation of predictive capacity, and even with external validation, no one can be sure whether the selection of training and test sets was manipulated to maximize the predictive capacity of the model being published. Different aspects of validation of QSAR models that need attention includes methods of selection of training set compounds,[14] setting training set size[15] and impact of variable selection[16] for training set models for determining the quality of prediction. Development of novel validation parameters for judging quality of QSAR models is also important.[17]
Application
Chemical
One of the first historical QSAR applications was to predict boiling points.[18]
It is well known for instance that within a particular family of chemical compounds, especially of organic chemistry, that there are strong correlations between structure and observed properties. A simple example is the relationship between the number of carbons in alkanes and their boiling points. There is a clear trend in the increase of boiling point with an increase in the number carbons and this serves as a means for predicting the boiling points of higher alkanes.
A still very interesting application is the Hammett equation, Taft equation and pKa prediction methods.[19]
Biological
The biological activity of molecules is usually measured in assays to establish the level of inhibition of particular signal transduction or metabolic pathways. Chemicals can also be biologically active by being toxic. Drug discovery often involves the use of QSAR to identify chemical structures that could have good inhibitory effects on specific targets and have low toxicity (non-specific activity). Of special interest is the prediction of partition coefficient log P, which is an important measure used in identifying "druglikeness" according to Lipinski's Rule of Five.
While many quantitative structure activity relationship analyses involve the interactions of a family of molecules with an enzyme or receptor binding site, QSAR can also be used to study the interactions between the structural domains of proteins. Protein-protein interactions can be quantitatively analyzed for structural variations resulted from site-directed mutagenesis.[20]
It is part of the machine learning method to reduce the risk for a SAR paradox, especially taking into account that only a finite amount of data is available (see also MVUE). In general all QSAR problems can be divided into a coding[21] and learning.[22]
Applicability domain
As the use of (Q)SAR models for chemical risk management increases steadily and is also used for regulatory purposes (in the EU: Registration, Evaluation, Authorisation and Restriction of Chemicals), it is of crucial importance to be able to assess the reliability of predictions. The chemical descriptor space spanned by a particular training set of chemicals is called Applicability Domain. It offers the opportunity to assess whether a compound can be reliably predicted.
See also
- ADME
- Cheminformatics
- Computer-assisted drug design (CADD)
- Conformation Activity Relationship
- Differential solubility
- Molecular design software
- Partition coefficient
- Pharmacokinetics
- Pharmacophore
- QSAR & Combinatorial Science – Scientific journal
- Software for molecular mechanics modeling
- Chemicalize.org:List of predicted structure based properties
References
- ^ Patani GA, LaVoie EJ (December 1996). "Bioisosterism: A Rational Approach in Drug Design". Chemical Reviews 96 (8): 3147–3176. doi:10.1021/cr950066q. PMID 11848856.
- ^ Wildman SA, Crippen GM (1999). "Prediction of physicochemical parameters by atomic contributions". J. Chem. Inf. Comput. Sci 39 (5): 868–873. doi:10.1021/ci990307l.
- ^ Ajmani S, Jadhav K, Kulkarni SA (November 2008). "Group-Based QSAR (G-QSAR): Mitigating Interpretation Challenges in QSAR". QSAR & Combinatorial Science 28 (1): 36–51. doi:10.1002/qsar.200810063.
- ^ Manoharan P, Vijayan RSK , Ghoshal N (September 2010). "Rationalizing fragment based drug discovery for BACE1: insights from FB-QSAR, FB-QSSR, multi objective (MO-QSPR) and MIF studies". Journal of Computer-Aided Molecular Design 24 (10): 843–864. doi:10.1007/s10822-010-9378-9. PMID 20740315.
- ^ Leach, Andrew R. (2001). Molecular modelling: principles and applications. Englewood Cliffs, N.J: Prentice Hall. ISBN 0-582-38210-6.
- ^ Vert, Jean-Philippe; Schl̲kopf, Bernhard; Schölkopf, Bernhard; Tsuda, Koji (2004). Kernel methods in computational biology. Cambridge, Mass: MIT Press. ISBN 0-262-19509-7.
- ^ Dietterich, Thomas G.; Lathrop, Richard H.; Lozano-Pérez, Tomás (1997). "Solving the multiple instance problem with axis-parallel rectangles". Artificial Intelligence 89 (1–2): 31–71. doi:10.1016/S0004-3702(96)00034-3.
- ^ Gusfield, Dan (1997). Algorithms on strings, trees, and sequences: computer science and computational biology. Cambridge, UK: Cambridge University Press. ISBN 0-521-58519-8.
- ^ Helma, Christoph (2005). Predictive toxicology. Washington, DC: Taylor & Francis. ISBN 0-8247-2397-X.
- ^ Tong W, Hong H, Xie Q, Shi L, Fang H, Perkins R (April 2005). "Assessing QSAR Limitations – A Regulatory Perspective". Current Computer-Aided Drug Design 1 (2): 195–205. doi:10.2174/1573409053585663. http://www.bentham.org/ccadd/contabs/cad1-2.htm#5.
- ^ Dearden JC (2003). "In silico prediction of drug toxicity". Journal of Computer-aided Molecular Design 17 (2–4): 119–27. doi:10.1023/A:1025361621494. PMID 13677480.
- ^ Wold S, Eriksson L (1995). "Statistical validation of QSAR results". In Waterbeemd, Han van de. Chemometric methods in molecular design. Weinheim: VCH. pp. 309–318. ISBN 3-527-30044-9.
- ^ Roy, K. (2007). "On some aspects of validation of predictive quantitative structure-activity relationship models". Expert Opin. Drug Discov. 2 (12): 1567–1577. doi:10.1517/17460441.2.12.1567.
- ^ Leonard JT, Roy K (2006). "On selection of training and test sets for the development of predictive QSAR models". QSAR & Combinatorial Science 25 (3): 235–251. doi:10.1002/qsar.200510161. http://library.sut.ac.th/free/ph_scie/pdfs/qsar/QSAR_3.pdf.
- ^ Roy PP, Leonard JT, Roy K (2008). "Exploring the impact of size of training sets for the development of predictive QSAR models". Chemometrics and Intelligent Laboratory Systems 90 (1): 31–42. doi:10.1016/j.chemolab.2007.07.004.
- ^ Roy PP, Roy K (2008). "On some aspects of variable selection for partial least squares regression models". QSAR & Combinatorial Science 27 (3): 302–313. doi:10.1002/qsar.200710043.
- ^ Roy PP, Paul S, Mitra I, Roy K (April 2009). "On Two Novel Parameters for Validation of Predictive QSAR Models". Molecules 14 (5): 1660–1701. doi:10.3390/molecules14051660. PMID 19471190. http://www.mdpi.com/1420-3049/14/5/1660/pdf.
- ^ Rouvray, D. H.; Bonchev, Danail (1991). Chemical graph theory: introduction and fundamentals. Tunbridge Wells, Kent, England: Abacus Press. ISBN 0-85626-454-7.
- ^ Fraczkiewicz R (2007). "In Silico Prediction of Ionization". Comprehensive medicinal chemistry II. Amsterdam: Elsevier. ISBN 0-08-044518-7.
- ^ Freyhult EK, Andersson K, Gustafsson MG (April 2003). "Structural Modeling Extends QSAR Analysis of Antibody-Lysozyme Interactions to 3D-QSAR". Biophysical Journal 84 (4): 2264–72. Bibcode 2003BpJ....84.2264F. doi:10.1016/S0006-3495(03)75032-2. PMC 1302793. PMID 12668435. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pmcentrez&artid=1302793.
- ^ Hendrik Timmerman; Todeschini, Roberto; Viviana Consonni; Raimund Mannhold; Hugo Kubinyi (2002). Handbook of Molecular Descriptors. Weinheim: Wiley-VCH. ISBN 3-527-29913-0.
- ^ Strok, David G.; Duda, Richard O.; Hart, Peter W. (2001). Pattern classification. Chichester: John Wiley & Sons. ISBN 0-471-05669-3.
Further reading
- Selassie CD (2003). "History of Quantitative Structure-Activity Relationships". In Abraham DJ. Burger's medicinal Chemistry and Drug Discovery. 1 (6th ed.). New York: Wiley. pp. 1–48. ISBN 0-471-27401-1. http://media.wiley.com/product_data/excerpt/03/04712709/0471270903.pdf.
External links
- "The Cheminformatics and QSAR Society". http://www.qsar.org/. Retrieved 2009-05-11.
- "The 3D QSAR Server". http://www.3d-qsar.com/. Retrieved 2011-06-18.
- "Nature Protocols: Development of QSAR models using C-QSAR program". Nature Protocols. doi:10.1038/nprot.2007.125. http://www.natureprotocols.com/2007/03/05/development_of_qsar_models_usi_1.php. Retrieved 2009-05-11. "A regression program that has dual databases of over 21,000 QSAR models"
- "QSAR World". http://www.qsarworld.com. Retrieved 2009-05-11. "A comprehensive web resource for QSAR modelers"
|
|||||