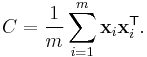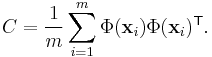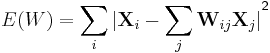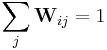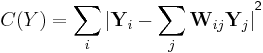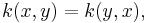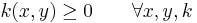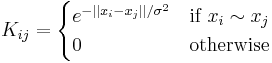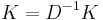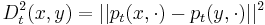Nonlinear dimensionality reduction
High-dimensional data, meaning data that requires more than two or three dimensions to represent, can be difficult to interpret. One approach to simplification is to assume that the data of interest lies on an embedded non-linear manifold within the higher-dimensional space. If the manifold is of low enough dimension then the data can be visualised in the low dimensional space.
Below is a summary of some of the important algorithms from the history of manifold learning and nonlinear dimensionality reduction.[1] Many of these non-linear dimensionality reduction methods are related to the linear methods listed below. Non-linear methods can be broadly classified into two groups: those that provide a mapping (either from the high dimensional space to the low dimensional embedding or vice versa), and those that just give a visualisation. In the context of machine learning, mapping methods may be viewed as a preliminary feature extraction step, after which pattern recognition algorithms are applied. Typically those that just give a visualisation are based on proximity data – that is, distance measurements.
Linear methods
- Independent component analysis (ICA).
- Principal component analysis (PCA) (also called Karhunen–Loève transform — KLT).
- Singular value decomposition (SVD).
- Factor analysis.
Uses for NLDR
Consider a dataset represented as a matrix (or a database table), such that each row represents a set of attributes (or features or dimensions) that describe a particular instance of something. If the number of attributes is large, then the space of unique possible rows is exponentially large. Thus, the larger the dimensionality, the more difficult it becomes to sample the space. This causes many problems. Algorithms that operate on high-dimensional data tend to have a very high time complexity. Many machine learning algorithms, for example, struggle with high-dimensional data. This has become known as the curse of dimensionality. Reducing data into fewer dimensions often makes analysis algorithms more efficient, and can help machine learning algorithms make more accurate predictions.
Humans often have difficulty comprehending data in many dimensions. Thus, reducing data to a small number of dimensions is useful for visualization purposes.
The reduced-dimensional representations of data are often referred to as "intrinsic variables". This description implies that these are the values from which the data was produced. For example, consider a dataset that contains images of a letter 'A', which has been scaled and rotated by varying amounts. Each image has 32x32 pixels. Each image can be represented as a vector of 1024 pixel values. Each row is a sample on a two-dimensional manifold in 1024-dimensional space (a Hamming space). The intrinsic dimensionality is two, because two variables (rotation and scale) were varied in order to produce the data. Information about the shape or look of a letter 'A' is not part of the intrinsic variables because it is the same in every instance. Nonlinear dimensionality reduction will discard the correlated information (the letter 'A') and recover only the varying information (rotation and scale). The image to the left shows sample images from this dataset (to save space, not all input images are shown), and a plot of the two-dimensional points that results from using a NLDR algorithm (in this case, Manifold Sculpting was used) to reduce the data into just two dimensions.
By comparison, if PCA (a linear dimensionality reduction algorithm) is used to reduce this same dataset into two dimensions, the resulting values are not so well organized. This demonstrates that the high-dimensional vectors (each representing a letter 'A') that sample this manifold vary in a non-linear manner.
It should be apparent, therefore, that NLDR has several applications in the field of computer-vision. For example, consider a robot that uses a camera to navigate in a closed static environment. The images obtained by that camera can be considered to be samples on a manifold in high-dimensional space, and the intrinsic variables of that manifold will represent the robot's position and orientation. This utility is not limited to robots. Dynamical systems, a more general class of systems, which includes robots, are defined in terms of a manifold. Active research in NLDR seeks to unfold the observation manifolds associated dynamical systems to develop techniques for modeling such systems and enable them to operate autonomously[2].
Manifold learning algorithms
Some of the more prominent manifold learning algorithms are listed below (in approximately chronological order). An algorithm may learn an internal model of the data, which can be used to map points unavailable at training time into the embedding in a process often called out-of-sample extension.
Sammon's mapping
Sammon's mapping is one of the first NLDR techniques.
Kohonen maps
Kohonen maps (also called self-organizing maps or SOM) and its probabilistic variant generative topographic mapping (GTM) use a point representation in the embedded space to form a latent variable model based on a non-linear mapping from the embedded space to the high dimensional space. These techniques are related to work on density networks, which also are based around the same probabilistic model.
Principal curves and manifolds
Principal curves and manifolds give the natural geometric framework for nonlinear dimensionality reduction and extend the geometric interpretation of PCA by explicitly constructing an embedded manifold, and by encoding using standard geometric projection onto the manifold. This approach was proposed by Trevor Hastie in his thesis (1984)[4] and developed further by many authors.[5] How to define the "simplicity" of the manifold is problem-dependent, however, it is commonly measured by the intrinsic dimensionality and/or the smoothness of the manifold. Usually, the principal manifold is defined as a solution to an optimization problem. The objective function includes a quality of data approximation and some penalty terms for the bending of the manifold. The popular initial approximations are generated by linear PCA, Kohonen's SOM or autoencoders. The elastic map method provides the expectation-maximization algorithm for principal manifold learning with minimization of quadratic energy functional at the "maximization" step.
Autoencoders
An autoencoder is a feed-forward neural network which is trained to approximate the identity function. That is, it is trained to map from a vector of values to the same vector. One of the hidden layers in the network, however, is limited to contain only a small number of network units. Thus, the network must learn to encode the vector into a small number of dimensions and then decode it back into the original space. Thus, the first half of the network is a model which maps from high to low-dimensional space, and the second half maps from low to high-dimensional space. Although the idea of autoencoders is quite old, training of the encoders has only recently become possible through the use of Restricted Boltzmann machines. Related to autoencoders is the NeuroScale algorithm, which uses stress functions inspired by multidimensional scaling and Sammon mappings (see below) to learn a non-linear mapping from the high dimensional to the embedded space. The mappings in NeuroScale are based on radial basis function networks.
Gaussian process latent variable models
Gaussian process latent variable models (GPLVM)[6] are a probabilistic non-linear PCA. Like kernel PCA they use a kernel function to form the mapping (in the form of a Gaussian process). However in the GPLVM the mapping is from the embedded space to the data space (like density networks and GTM) whereas in kernel PCA it is in the opposite direction.
Curvilinear component analysis
Curvilinear component analysis (CCA) [7] looks for the configuration of points in the output space that preserves original distances as much as possible while focusing on small distances in the output space (conversely to Sammon's mapping which focus on small distances in original space).
It should be noticed that CCA, as an iterative learning algorithm, actually starts with focus on large distances (like the Sammon algorithm), then gradually change focus to small distances. The small distance information will overwrite the large distance information, if compromises between the two have to be made.
The stress function of CCA has been proved as a sum of right Bregman divergence [8]
Curvilinear distance analysis
CDA[7] trains a self-organizing neural network to fit the manifold and seeks to preserve geodesic distances in its embedding. It is based on Curvilinear Component Analysis (which extended Sammon's mapping), but uses geodesic distances instead.
Diffeomorphic dimensionality reduction
Diffeomorphic Dimensionality Reduction or Diffeomap[9] learns a smooth diffeomorphic mapping which transports the data onto a lower dimensional linear subspace. The methods solves for a smooth time indexed vector field such that path integrals along the field which start at the data points will end at a lower dimensional linear subspace, thereby attempting to preserve pairwise differences under both the forward and inverse mapping.
Kernel principal component analysis
Perhaps the most widely used algorithm for manifold learning is kernel PCA.[10] It is a combination of Principal component analysis and the kernel trick. PCA begins by computing the covariance matrix of the 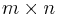 matrix
matrix 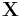
It then projects the data onto the first k eigenvectors of that matrix. By comparison, KPCA begins by computing the covariance matrix of the data after being transformed into a higher-dimensional space,
It then projects the transformed data onto the first k eigenvectors of that matrix, just like PCA. It uses the kernel trick to factor away much of the computation, such that the entire process can be performed without actually computing 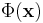 . Of course
. Of course 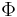 must be chosen such that it has a known corresponding kernel. Unfortunately, it is not trivial to find a good kernel for a given problem, so KPCA does not yield good results with some problems. For example, it is known to perform poorly with the swiss roll manifold.
must be chosen such that it has a known corresponding kernel. Unfortunately, it is not trivial to find a good kernel for a given problem, so KPCA does not yield good results with some problems. For example, it is known to perform poorly with the swiss roll manifold.
KPCA has an internal model, so it can be used to map points onto its embedding that were not available at training time.
Isomap
Isomap[11] is a combination of the Floyd–Warshall algorithm with classic Multidimensional Scaling. Classic Multidimensional Scaling (MDS) takes a matrix of pair-wise distances between all points, and computes a position for each point. With NLDR algorithms like Isomap, however, the pair-wise distances are only known between neighboring points. So Isomap uses the Floyd–Warshall algorithm to compute the pair-wise distances between all of the other points. This effectively estimates the full matrix of pair-wise geodesic distances between all of the points. Isomap then uses classic MDS to compute the reduced-dimensional positions of all the points.
Landmark-Isomap is a variant of this algorithm that uses landmarks to increase speed, at the cost of some accuracy.
Locally-linear embedding
Locally-Linear Embedding (LLE)[12] was presented at approximately the same time as Isomap. It has several advantages over Isomap, including faster optimization when implemented to take advantage of sparse matrix algorithms, and better results with many problems. LLE also begins by finding a set of the nearest neighbors of each point. It then computes a set of weights for each point that best describe the point as a linear combination of its neighbors. Finally, it uses an eigenvector-based optimization technique to find the low-dimensional embedding of points, such that each point is still described with the same linear combination of its neighbors. LLE tends to handle non-uniform sample densities poorly because there is no fixed unit to prevent the weights from drifting as various regions differ in sample densities. LLE has no internal model.
LLE computes the barycentric coordinates of a point Xi based on its neighbors Xj. The original point is reconstructed by a linear combination, given by the weight matrix Wij, of its neighbors. The reconstruction error is given by the cost function E(W).
The weights Wij refer to the amount of contribution the point Xj has while reconstructing the point Xi. The cost function is minimized under two constraints: (a) Each data point Xi is reconstructed only from its neighbors, thus enforcing Wij to be zero if point Xj is not a neighbor of the point Xi and (b) The sum of every row of the weight matrix equals 1.
The original data points are collected in a D dimensional space and the goal of the algorithm is to reduce the dimensionality to d such that D >> d. The same weights Wij that reconstructs the ith data point in the D dimensional space will be used to reconstruct the same point in the lower d dimensional space. A neighborhood preserving map is created based on this idea. Each point Xi in the D dimensional space is mapped onto a point Yi in the d dimensional space by minimizing the cost function
In this cost function unlike the previous one the weights Wij are kept fixed and the minimization is done on the points Yi to optimize the coordinates. This minimization problem can be solved by solving a sparse N X N eigen value problem, whose bottom d nonzero eigen vectors provide an orthogonal set of coordinates. Generally the data points are reconstructed from K nearest neighbors, as measured by Euclidean distance. For such an implementation the algorithm has only one free parameter K, which can be chosen by cross validation.
Laplacian eigenmaps
Laplacian Eigenmaps [13] uses spectral techniques to perform dimensionality reduction. This technique relies on the basic assumption that the data lies in a low dimensional manifold in a high dimensional space.[14] This algorithm cannot embed out of sample points, but techniques based on Reproducing kernel Hilbert space regularization exist for adding this capability.[15] Such techniques can be applied to other nonlinear dimensionality reduction algorithms as well.
Traditional techniques like principal component analysis do not consider the intrinsic geometry of the data. Laplacian eigenmaps builds a graph from neighborhood information of the data set. Each data point serves as a node on the graph and connectivity between nodes is governed by the proximity of neighboring points (using e.g. the k-nearest neighbor algorithm). The graph thus generated can be considered as a discrete approximation of the low dimensional manifold in the high dimensional space. Minimization of a cost function based on the graph ensures that points close to each other on the manifold are mapped close to each other in the low dimensional space, preserving local distances. The eigenfunctions of the Laplace–Beltrami operator on the manifold serve as the embedding dimensions, since under mild conditions this operator has a countable spectrum that is a basis for square integrable functions on the manifold (compare to Fourier series on the unit circle manifold). Attempts to place Laplacian eigenmaps on solid theoretical ground have met with some success, as under certain nonrestrictive assumptions, the graph Laplacian matrix has been shown to converge to the Laplace–Beltrami operator as the number of points goes to infinity.[16] Matlab code for Laplacian Eigenmaps can be found in algorithms[17] and the PhD thesis of Belkin can be found at the Ohio State University.[18]
Diffusion maps
Diffusion maps leverages the relationship between heat diffusion and a random walk (Markov Chain); an analogy is drawn between the diffusion operator on a manifold and a Markov transition matrix operating on functions defined on the graph whose nodes were sampled from the manifold.[19]. In particular let a data set be represented by ![\mathbf{X} = [x_1,x_2,\ldots,x_n] \in \Omega \subset \mathbf {R^D}](/2012-wikipedia_en_all_nopic_01_2012/I/4da5ef48242bf13252e2f3340d5028fc.png) . The underlying assumption of diffusion map is that the data although high-dimensional, lies on a low-dimensional manifold of dimensions
. The underlying assumption of diffusion map is that the data although high-dimensional, lies on a low-dimensional manifold of dimensions 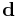 .X represents the data set and let
.X represents the data set and let 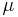 represent the distribution of the data points on X. In addition to this lets define a kernel which represents some notion of affinity of the points in X. The kernel
represent the distribution of the data points on X. In addition to this lets define a kernel which represents some notion of affinity of the points in X. The kernel 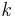 has the following properties [20]
has the following properties [20]
k is symmetric
k is positivity preserving
Thus one can think of the individual data points as the nodes of a graph and the kernel k defining some sort of affinity on that graph. The graph is symmetric by construction since the kernel is symmetric. It is easy to see here that from the tuple {X,k} one can construct a reversible Markov Chain. This technique is fairly popular in a variety of fields and is known as the graph laplacian.
The graph K = (X,E) can be constructed for example using a Gaussian kernel.
In this above equation 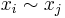 denotes that
denotes that 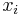 is a nearest neighbor of
is a nearest neighbor of 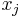 . In reality Geodesic distance should be used to actually measure distances on the manifold. Since the exact structure of the manifold is not available, the geodesic distance is approximated by euclidean distances with only nearest neighbors. The choice
. In reality Geodesic distance should be used to actually measure distances on the manifold. Since the exact structure of the manifold is not available, the geodesic distance is approximated by euclidean distances with only nearest neighbors. The choice 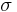 modulates our notion of proximity in the sense that if
modulates our notion of proximity in the sense that if 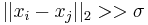 then
then 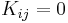 and if
and if 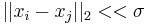 then
then 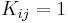 . The former means that very little diffusion has taken place while the latter implies that the diffusion process is nearly complete. Different strategies to choose
. The former means that very little diffusion has taken place while the latter implies that the diffusion process is nearly complete. Different strategies to choose 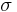 can be found in [21]. If
can be found in [21]. If 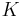 has to faithfully represent a Markov Matrix, then it has to be normalized by the corresponding degree matrix
has to faithfully represent a Markov Matrix, then it has to be normalized by the corresponding degree matrix 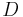 .
.
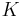 now represents a Markov chain.
now represents a Markov chain. 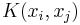 is the probability of transitioning from
is the probability of transitioning from 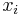 to
to 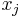 in one a time step. Similarly the probability of transitioning from
in one a time step. Similarly the probability of transitioning from 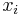 to
to 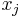 in t time steps is given by
in t time steps is given by 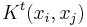 . Here
. Here 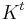 is the matrix K multiplied to itself t times. Now the kernel markov matrix K constitutes some notion of local geometry of the data set X. The major difference between diffusion maps and principal component analysis is that only local features of the data is considered in diffusion maps as opposed to taking correlations of the entire data set.
is the matrix K multiplied to itself t times. Now the kernel markov matrix K constitutes some notion of local geometry of the data set X. The major difference between diffusion maps and principal component analysis is that only local features of the data is considered in diffusion maps as opposed to taking correlations of the entire data set.
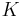 defines a random walk on the data set which means that the kernel captures some local geometry of data set. The markov chain defines fast and slow directions of propagation , based on the values taken by the kernel, and as one propagates the walk forward in time, the local geometry information is being aggregated in the same way as local transitions (defined by differential equations) of the dynamical system [22]. The concept of diffusion arises from the definition of a family diffusion distance {
defines a random walk on the data set which means that the kernel captures some local geometry of data set. The markov chain defines fast and slow directions of propagation , based on the values taken by the kernel, and as one propagates the walk forward in time, the local geometry information is being aggregated in the same way as local transitions (defined by differential equations) of the dynamical system [22]. The concept of diffusion arises from the definition of a family diffusion distance {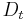 }
}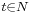
For a given value of t 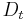 defines a distance between any two points of the data set. This means that the value of
defines a distance between any two points of the data set. This means that the value of 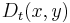 will be small if there are many paths that connect x to y and vice versa. The quantity
will be small if there are many paths that connect x to y and vice versa. The quantity 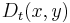 involves summing over of all paths of length t, as a result of which
involves summing over of all paths of length t, as a result of which 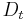 is extremely robust to noise in the data as opposed to geodesic distance.
is extremely robust to noise in the data as opposed to geodesic distance. 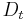 takes into account all the relation between points x and y while calculating the distance and serves as a better notion of proximity than just Euclidean distance or even geodesic distance.
takes into account all the relation between points x and y while calculating the distance and serves as a better notion of proximity than just Euclidean distance or even geodesic distance.
Hessian LLE
Like LLE, Hessian LLE [23] is also based on sparse matrix techniques. It tends to yield results of a much higher quality than LLE. Unfortunately, it has a very costly computational complexity, so it is not well-suited for heavily-sampled manifolds. It has no internal model.
Modified LLE
Modified LLE (MLLE) [24] is another LLE variant which uses multiple weights in each neighborhood to address the local weight matrix conditioning problem which leads to distortions in LLE maps. MLLE produces robust projections similar to Hessian LLE, but without the significant additional computational cost.
Local tangent space alignment
LTSA[25] is based on the intuition that when a manifold is correctly unfolded, all of the tangent hyperplanes to the manifold will become aligned. It begins by computing the k-nearest neighbors of every point. It computes the tangent space at every point by computing the d-first principal components in each local neighborhood. It then optimizes to find an embedding that aligns the tangent spaces.
Local multidimensional scaling
Local Multidimensional Scaling[26] performs multidimensional scaling in local regions, and then uses convex optimization to fit all the pieces together.
Maximum variance unfolding
Maximum Variance Unfolding was formerly known as Semidefinite Embedding. The intuition for this algorithm is that when a manifold is properly unfolded, the variance over the points is maximized. This algorithm also begins by finding the k-nearest neighbors of every point. It then seeks to solve the problem of maximizing the distance between all non-neighboring points, constrained such that the distances between neighboring points are preserved. The primary contribution of this algorithm is a technique for casting this problem as a semidefinite programming problem. Unfortunately, semidefinite programming solvers have a high computational cost. The Landmark–MVU variant of this algorithm uses landmarks to increase speed with some cost to accuracy. It has no model.
Nonlinear PCA
Nonlinear PCA[27] (NLPCA) uses backpropagation to train a multi-layer perceptron to fit to a manifold. Unlike typical MLP training, which only updates the weights, NLPCA updates both the weights and the inputs. That is, both the weights and inputs are treated as latent values. After training, the latent inputs are a low-dimensional representation of the observed vectors, and the MLP maps from that low-dimensional representation to the high-dimensional observation space.
Data-driven high-dimensional scaling
Data-Driven High Dimensional Scaling (DD-HDS) [28] is closely related to Sammon's mapping and curvilinear component analysis except that (1) it simultaneously penalizes false neighborhoods and tears by focusing on small distances in both original and output space, and that (2) it accounts for concentration of measure phenomenon by adapting the weighting function to the distance distribution. Data-Driven High Dimensional Scaling (DD-HDS) [29] is closely related to Sammon's mapping and curvilinear component analysis except that (1) it simultaneously penalizes false neighborhoods and tears by focusing on small distances in both original and output space, and that (2) it accounts for concentration of measure phenomenon by adapting the weighting function to the distance distribution.
Manifold sculpting
Manifold Sculpting[30] uses graduated optimization to find an embedding. Like other algorithms, it computes the k-nearest neighbors and tries to seek an embedding that preserves relationships in local neighborhoods. It slowly scales variance out of higher dimensions, while simultaneously adjusting points in lower dimensions to preserve those relationships. If the rate of scaling is small, it can find very precise embeddings. It boasts higher empirical accuracy than other algorithms with several problems. It can also be used to refine the results from other manifold learning algorithms. It struggles to unfold some manifolds, however, unless a very slow scaling rate is used. It has no model.
RankVisu
RankVisu [31] is designed to preserve rank of neighborhood rather than distance. RankVisu is especially useful on difficult tasks (when the preservation of distance cannot be achieved satisfyingly). Indeed, the rank of neighborhood is less informative than distance (ranks can be deduced from distances but distances cannot be deduced from ranks) and its preservation is thus easier.
Topologically constrained isometric embedding
Topologically Constrained Isometric Embedding (TCIE) [32] is an algorithm based approximating geodesic distances after filtering geodesics inconsistent with the Euclidean metric. Aimed at correcting the distortions caused when Isomap is used to map intrinsically non-convex data, TCIE uses weight least-squares MDS in order to obtain a more accurate mapping. The TCIE algorithm first detects possible boundary points in the data, and during computation of the geodesic length marks inconsistent geodesics, to be given a small weight in the weighted Stress majorization that follows.
Relational perspective map
Relational perspective map is a special class of multidimensional scaling algorithm. The algorithm finds a configuration of data points on a manifold by simulating a multi-particle dynamic system on a closed manifold, where data points are mapped to particles and distances (or dissimilarity) between data points are used as strength of a kind of repulsive force. As the manifold gradually grows in size the multi-particale system cools down gradually and converges to configuration that reflects the distance information of the data points.
Relational perspective map is initially inspired by the physic system in which positively charged particles freely move on the surface of a ball. Guided by Coulumbian repulsive force between particles, the minimal energy configuration of the particles will reflect the strength of repulsive forces between the particles.
Relational perspective map is introduced in.[33] The algorithm firstly used the flat torus as the image manifold, then it has been extended (in the software VisuMap to use other types of closed manifolds, like the sphere, projective space, and Klein bottle, as image manifolds.
Methods based on proximity matrices
A method based on proximity matrices is one where the data is presented to the algorithm in the form of a similarity matrix or a distance matrix. These methods all fall under the broader class of metric multidimensional scaling. The variations tend to be differences in how the proximity data is computed; for example, Isomap, locally linear embeddings, maximum variance unfolding, and Sammon mapping (which is not in fact a mapping) are examples of metric multidimensional scaling methods.
See also
- Discriminant analysis
- Elastic map[34]
- Pairwise distance methods
- Self-organizing map (SOM)
- Topographic map
References
- ^ John A. Lee, Michel Verleysen, Nonlinear Dimensionality Reduction, Springer, 2007.
- ^ Gashler, M. and Martinez, T., Temporal Nonlinear Dimensionality Reduction, In Proceedings of the International Joint Conference on Neural Networks IJCNN'11, pp. 1959–1966, 2011
- ^ A. N. Gorban, A. Zinovyev, Principal manifolds and graphs in practice: from molecular biology to dynamical systems, International Journal of Neural Systems, Vol. 20, No. 3 (2010) 219–232.
- ^ T. Hastie, Principal Curves and Surfaces, Ph.D Dissertation, Stanford Linear Accelerator Center, Stanford University, Stanford, California, US, November 1984.
- ^ A. Gorban, B. Kegl, D. Wunsch, A. Zinovyev (Eds.), Principal Manifolds for Data Visualisation and Dimension Reduction, LNCSE 58, Springer, Berlin – Heidelberg – New York, 2007. ISBN 978-3-540-73749-0
- ^ N. Lawrence, Probabilistic Non-linear Principal Component Analysis with Gaussian Process Latent Variable Models, Journal of Machine Learning Research 6(Nov):1783–1816, 2005.
- ^ a b P. Demartines and J. Hérault, Curvilinear Component Analysis: A Self-Organizing Neural Network for Nonlinear Mapping of Data Sets, IEEE Transactions on Neural Networks, Vol. 8(1), 1997, pp. 148–154
- ^ Jigang Sun, Malcolm Crowe, and Colin Fyfe, Curvilinear component analysis and Bregman divergences, In European Symposium on Artificial Neural Networks (Esann), pages 81–86. d-side publications, 2010
- ^ Christian Walder and Bernhard Schölkopf, Diffeomorphic Dimensionality Reduction, Advances in Neural Information Processing Systems 22, 2009, pp. 1713–1720, MIT Press
- ^ B. Schölkopf, A. Smola, K.-R. Muller, Kernel Principal Component Analysis, In: Bernhard Schölkopf, Christopher J. C. Burges, Alexander J. Smola (Eds.), Advances in Kernel Methods-Support Vector Learning, 1999, MIT Press Cambridge, MA, USA, 327–352. ISBN 0-262-19416-3
- ^ J. B. Tenenbaum, V. de Silva, J. C. Langford, A Global Geometric Framework for Nonlinear Dimensionality Reduction, Science 290, (2000), 2319–2323.
- ^ S. T. Roweis and L. K. Saul, Nonlinear Dimensionality Reduction by Locally Linear Embedding, Science Vol 290, 22 December 2000, 2323–2326.
- ^ Mikhail Belkin and Partha Niyogi, Laplacian Eigenmaps and Spectral Techniques for Embedding and Clustering, Advances in Neural Information Processing Systems 14, 2001, p. 586–691, MIT Press
- ^ Mikhail Belkin Problems of Learning on Manifolds, PhD Thesis, Department of Mathematics, The University Of Chicago, August 2003
- ^ Bengio et al. "Out-of-Sample Extensions for LLE, Isomap, MDS, Eigenmaps, and Spectral Clustering" in Advances in Neural Information Processing Systems (2004)
- ^ Mikhail Belkin Problems of Learning on Manifolds, PhD Thesis, Department of Mathematics, The University Of Chicago, August 2003
- ^ Ohio-state.edu
- ^ Ohio-state.edu
- ^ Diffusion Maps and Geometric Harmonics, Stephane Lafon, PhD Thesis, Yale University, May 2004
- ^ Diffusion Maps, Ronald R. Coifman and Stephane Lafon,: Science, 19 June, 2006
- ^ B. Bah, "Diffusion Maps: Applications and Analysis", Masters Thesis, University of Oxford
- ^ Diffusion Maps, Ronald R. Coifman and Stephane Lafon,: Science, 19 June , 2006
- ^ D. Donoho and C. Grimes, "Hessian eigenmaps: Locally linear embedding techniques for high-dimensional data" Proc Natl Acad Sci U S A. 2003 May 13; 100(10): 5591–5596
- ^ Z. Zhang and J. Wang, "MLLE: Modified Locally Linear Embedding Using Multiple Weights" http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.70.382
- ^ Zhang, Zhenyue; Hongyuan Zha (2005). "Principal Manifolds and Nonlinear Dimension Reduction via Local Tangent Space Alignment". SIAM Journal on Scientific Computing 26 (1): 313–338.
- ^ J Venna and S Kaski, Local multidimensional scaling, Neural Networks, 2006
- ^ Scholz, M. Kaplan, F. Guy, C. L. Kopka, J. Selbig, J., Non-linear PCA: a missing data approach, In Bioinformatics, Vol. 21, Number 20, pp. 3887–3895, Oxford University Press, 2005
- ^ S. Lespinats, M. Verleysen, A. Giron, B. Fertil, DD-HDS: a tool for visualization and exploration of high dimensional data, IEEE Transactions on Neural Networks 18 (5) (2007) 1265–1279.
- ^ S. Lespinats, M. Verleysen, A. Giron, B. Fertil, DD-HDS: a tool for visualization and exploration of high dimensional data, IEEE Transactions on Neural Networks 18 (5) (2007) 1265–1279.
- ^ Gashler, M. and Ventura, D. and Martinez, T., Iterative Non-linear Dimensionality Reduction with Manifold Sculpting, In Platt, J.C. and Koller, D. and Singer, Y. and Roweis, S., editor, Advances in Neural Information Processing Systems 20, pp. 513–520, MIT Press, Cambridge, MA, 2008
- ^ Lespinats S., Fertil B., Villemain P. and Herault J., Rankvisu: Mapping from the neighbourhood network, Neurocomputing, vol. 72 (13–15), pp. 2964–2978, 2009.
- ^ Rosman G., Bronstein M. M., Bronstein A. M. and Kimmel R., Nonlinear Dimensionality Reduction by Topologically Constrained Isometric Embedding, International Journal of Computer Vision, Volume 89, Number 1, 56–68, 2010
- ^ James X. Li, Visualizing high dimensional data with relational perspective map, Information Visualization (2004) 3, 49–59
- ^ ELastic MAPs
External links
- Isomap
- Generative Topographic Mapping
- Mike Tipping's Thesis
- Gaussian Process Latent Variable Model
- Locally Linear Embedding
- Relational Perspective Map
- Waffles is an open source C++ library containing implementations of LLE, Manifold Sculpting, and some other manifold learning algorithms.
- DD-HDS homepage
- RankVisu homepage
- Short review of Diffusion Maps