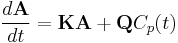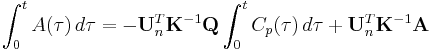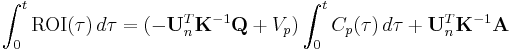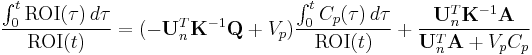Logan plot
A Logan plot (or Logan graphical analysis)[1] is a graphical analysis technique based on the compartment model that uses linear regression to analyze pharmacokinetics of tracers involving reversible uptake. It is mainly used for the evaluation of nuclear medicine imaging data after the injection of a labeled ligand that binds reversibly to specific receptor or enzyme.
In conventional compartmental analysis, an iterative method is used to fit the individual model parameters in the solution of a compartmental model of specific configuration to the measurements with a measured plasma time-activity curve that serves as an forcing (input) function, and the binding of the tracer can then be described. Graphical analysis is a simplified method that transforms the model equations into a linear equation evaluated at multiple time points and provides fewer parameters (i.e., slope and intercept). Although the slope and the intercept can be interpreted in terms of a combination of model parameters if a compartmental model configuration is assumed, the graphical methods are independent of any specific model configuration. In case of irreversible tracers, certain fraction of the radioactivity is trapped in the tissue or the binding site during the course of the experiment, whereas reversible tracers show uptake and loss from all compartments throughout the study. The theoretical foundation of graphical analysis for irreversible tracers (also called Patlak graphical analysis or Patlak plot) was laid by Clifford Patlak and his colleagues[2][3] at NIH. Based on the original work of Patlak, Jean Logan and her colleagues[1] from Brookhaven National Laboratory extended the method to tracers with reversible kinetics.
The kinetics of radiolabeled compounds in a compartmental system can be described in terms of a set of first-order, constant-coefficient, ordinary differential equations.[4][5] The time course of the activity in the multicompartmental system driven by a metabolite-corrected plasma input function 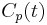 can be described by:
can be described by:
where 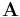 is a column vector of activity concentration for each compartment at time
is a column vector of activity concentration for each compartment at time 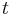 ,
, 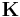 is the matrix of the transfer constants between compartments, and
is the matrix of the transfer constants between compartments, and 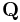 is the vector of plasma-to-tissue transfer constants. Patlak and Blasberg[3] showed that the above equation can be written as:
is the vector of plasma-to-tissue transfer constants. Patlak and Blasberg[3] showed that the above equation can be written as:
where 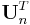 represents a row vector of 1s and
represents a row vector of 1s and 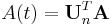 . The total activity in the region of interest,
. The total activity in the region of interest, 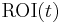 , is a combination of radioactivities from all compartments plus a plasma volume fraction (
, is a combination of radioactivities from all compartments plus a plasma volume fraction ( )[2] and thus:
)[2] and thus:
By dividing both sides by 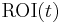 , one obtains the following linear equation:
, one obtains the following linear equation:
For 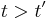 , Patlak and his colleagues[2] showed that
, Patlak and his colleagues[2] showed that 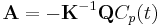 , i.e., the steady-state condition. When this condition is satisfied, the intercept has reached its constant value so that after some time a plot of
, i.e., the steady-state condition. When this condition is satisfied, the intercept has reached its constant value so that after some time a plot of 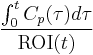 versus
versus 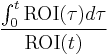 becomes a straight line with slope
becomes a straight line with slope 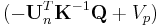 and intercept
and intercept 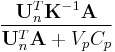 .[1]
.[1]
For a catenary two-tissue compartment model with transfer constants 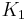 (forward transport from plasma to tissue),
(forward transport from plasma to tissue), 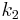 (reverse transport from tissue to plasma),
(reverse transport from tissue to plasma), 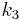 (binding parameter proportional to
(binding parameter proportional to 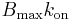 ), and
), and 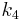 (dissociation constant) to analyze enzyme or receptor system, the slope represents the total distribution volume (
(dissociation constant) to analyze enzyme or receptor system, the slope represents the total distribution volume (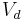 ) and is given by
) and is given by 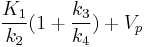 [1], where
[1], where 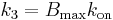 ,
, 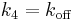 ,
, 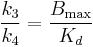 , and
, and 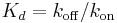 , in which
, in which 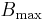 is the concentration of ligand binding sites,
is the concentration of ligand binding sites, 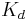 is the equilibrium dissociation constant for the ligand-binding site complex,
is the equilibrium dissociation constant for the ligand-binding site complex, 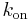 is the ligand-binding association constant,
is the ligand-binding association constant, 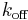 is the ligand-binding dissociation constant. For a one-tissue compartment model with transfer constants
is the ligand-binding dissociation constant. For a one-tissue compartment model with transfer constants 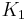 and
and 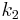 , the slope is
, the slope is 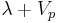 , where
, where 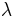 is the partition coefficient (
is the partition coefficient (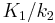 ) and the intercept is
) and the intercept is 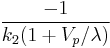 .[1]
.[1]
See also
- Patlak plot
- Multi-compartment model
- Positron emission tomography
- Binding potential
- Distribution volume
References
- ^ a b c d e J. Logan, J.S. Fowler, N.D. Volkow, A.P. Wolf, S.L. Dewey, D.J. Schlyer, R.R. MacGregor, R. Hitzemann, B. Bendriem, S.J. Gatley, D.R. Christman (September 1990). "Graphical analysis of reversible radioligand binding from time-activity measurements applied to [N-11C-methyl]-(-)-cocaine PET studies in human subjects". Journal of Cerebral Blood Flow and Metabolism 10 (5): 740–747. doi:10.1038/jcbfm.1990.127. PMID 2384545.
- ^ a b c C.S. Patlak, R.G. Blasberg, J.D. Fenstermacher (March 1983). "Graphical evaluation of blood-to-brain transfer constants from multiple-time uptake data". Journal of Cerebral Blood Flow and Metabolism 3 (1): 1–7. doi:10.1038/jcbfm.1983.1. PMID 6822610.
- ^ a b C.S. Patlak, R.G. Blasberg (April 1985). "Graphical evaluation of blood-to-brain transfer constants from multiple-time uptake data. Generalizations". Journal of Cerebral Blood Flow and Metabolism 5 (4): 584–590. doi:10.1038/jcbfm.1985.87. PMID 4055928.
- ^ K. Godfrey (1983). Compartmental Models and Their Application. Academic Press, New York.
- ^ J.A. Jacquez (1985). Compartmental Analysis in Biology and Medicine (2nd ed.). The University of Michigan Press, Ann Arbor.