Prion
| Prion Diseases (TSEs) | |
|---|---|
| Classification and external resources | |
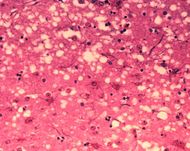 Microscopic "holes" are characteristic in prion-affected tissue sections, causing the tissue to develop a "spongy" architecture. |
|
| ICD-10 | A81 |
| ICD-9 | 046 |
A proteinaceous infectious particle, or prion, (pronounced /ˈpriː.ɒn/ (![]() listen)[1]) is an infectious agent composed primarily of protein.[2] The word prion, coined in 1982 by Dr. Stanley B. Prusiner, is a portmanteau derived from the words protein and infection.[3] Prions are the cause of a number of diseases in a variety of mammals, including bovine spongiform encephalopathy (BSE, also known as "mad cow disease") in cattle and Creutzfeldt–Jakob disease (CJD) in humans. In general usage, prion refers to the theoretical unit of infection. All known prion diseases affect the structure of the brain or other neural tissue and all are currently untreatable and universally fatal.[4]
listen)[1]) is an infectious agent composed primarily of protein.[2] The word prion, coined in 1982 by Dr. Stanley B. Prusiner, is a portmanteau derived from the words protein and infection.[3] Prions are the cause of a number of diseases in a variety of mammals, including bovine spongiform encephalopathy (BSE, also known as "mad cow disease") in cattle and Creutzfeldt–Jakob disease (CJD) in humans. In general usage, prion refers to the theoretical unit of infection. All known prion diseases affect the structure of the brain or other neural tissue and all are currently untreatable and universally fatal.[4]
Prions propagate by transmitting a mis-folded protein state: so as with viruses the protein cannot replicate by itself. Instead, when a prion enters a healthy organism the prion form of a protein induces pre-existing normal forms of the protein to convert into the rogue form. Since the new prions can then go on to convert more proteins themselves, this triggers a chain reaction that produces large amounts of the prion form.[5] All known prions induce the formation of an amyloid fold, in which the protein polymerises into an aggregate consisting of tightly packed beta sheets. Amyloid aggregates are fibrils, growing at their ends, and replicating when breakage causes two growing ends to become four growing ends. The incubation period of prion diseases is determined by the exponential growth rate associated with prion replication, which is a balance between the linear growth and the breakage of aggregates.[6]
This altered structure is extremely stable and accumulates in infected tissue, causing tissue damage and cell death.[7] This structural stability means that prions are resistant to denaturation by chemical and physical agents, making disposal and containment of these particles difficult. Prions come in different strains, each with a slightly different structure, and most of the time, strains breed true. Prion replication is nevertheless subject to occasional epimutation and then natural selection just like other forms of replication.[8] However, the number of possible distinct prion strains is likely far smaller than the number of possible DNA sequences, so evolution takes place within a limited space.
In scientific notation, PrPC refers to the endogenous form of prion protein (PrP), which is found in a multitude of tissues, while PrPSc refers to the misfolded form of PrP, that is responsible for the formation of amyloid plaques[9] and neurodegeneration.[10] The precise structure of the prion is not known, though they can be formed by combining PrPC, polyadenylic acid, and lipids.[11] Proteins showing prion-type behavior are also found in some fungi, which has been useful in helping to understand mammalian prions. Fungal prions, however, do not appear to cause disease in their hosts and may even confer an evolutionary advantage through a form of protein-based inheritance.[12]
Contents |
Discovery
Radiation biologist Tikvah Alper and mathematician John Stanley Griffith developed the hypothesis during the 1960s that some transmissible spongiform encephalopathies are caused by an infectious agent consisting solely of proteins.[13][14] Their theory was developed to explain the discovery that the mysterious infectious agent causing the diseases scrapie and Creutzfeldt–Jakob disease resisted ultraviolet radiation. Francis Crick recognized the potential importance of the Griffith protein-only hypothesis for scrapie propagation in the second edition of his "Central dogma of molecular biology": while asserting that the flow of sequence information from protein to protein, or from protein to RNA and DNA was "precluded". He noted that Griffith's hypothesis was a potential contradiction (although it was not so promoted by Griffith).[15] The revised hypothesis was later formulated, in part, to accommodate discovery of reverse transcription by Howard Temin and David Baltimore.
Stanley B. Prusiner of the University of California, San Francisco announced in 1982 that his team had purified the hypothetical infectious prion, and that the infectious agent consisted mainly of a specific protein – though they did not manage to isolate the protein until two years after Prusiner's announcement.[16] Prusiner coined the word "prion" as a name for the infectious agent. While the infectious agent was named a prion, the specific protein that the prion was composed of is also known as the Prion Protein (PrP), though this protein may occur both in infectious and non-infectious forms. Prusiner was awarded the Nobel Prize in Physiology or Medicine in 1997 for his research into prions.[17]
Structure
Isoforms
The protein that prions are made of (PrP) is found throughout the body, even in healthy people and animals. However, PrP found in infectious material has a different structure and is resistant to proteases, the enzymes in the body that can normally break down proteins. The normal form of the protein is called PrPC, while the infectious form is called PrPSc — the C refers to 'cellular' or 'common' PrP, while the Sc refers to 'scrapie', a prion disease occurring in sheep.[18] While PrPC is structurally well-defined, PrPSc is certainly polydisperse and defined at a relatively poor level. PrP can be induced to fold into other more-or-less well-defined isoforms in vitro, and their relationship to the form(s) that are pathogenic in vivo is not yet clear.
PrPC
PrPC is a normal protein found on the membranes of cells. It has 209 amino acids (in humans), one disulfide bond, a molecular weight of 35-36 kDa and a mainly alpha-helical structure. Several topological forms exist; one cell surface form anchored via glycolipid and two transmembrane forms.[19] The normal protein is not sedimentable; meaning it cannot be separated by centrifuging techniques.[9] Its function is a complex issue that continues to be investigated. PrPC binds copper (II) ions with high affinity.[20] The significance of this finding is not clear, but it presumably relates to PrP structure or function. PrPC is readily digested by proteinase K and can be liberated from the cell surface in vitro by the enzyme phosphoinositide phospholipase C (PI-PLC), which cleaves the glycophosphatidylinositol (GPI) glycolipid anchor.[21] PrP has been reported to play important roles in cell-cell adhesion and intracellular signaling in vivo, and may therefore be involved in cell-cell communication in the brain.[22]
PrPSc
The infectious isoform of PrP, known as PrPSc, is able to convert normal PrPC proteins into the infectious isoform by changing their conformation, or shape; this, in turn, alters the way the proteins interconnect. Although the exact 3D structure of PrPSc is not known, it has a higher proportion of β-sheet structure in place of the normal α-helix structure.[23] Aggregations of these abnormal isoforms form highly structured amyloid fibers, which accumulate to form plaques. It is unclear if these aggregates are the cause of cell damage or are simply a side effect of the underlying disease process.[24] The end of each fiber acts as a template onto which free protein molecules may attach, allowing the fiber to grow. Only PrP molecules with an identical amino acid sequence to the infectious PrPSc are incorporated into the growing fiber.[9] Although this property is not strictly shared by other proteins considered prions. The sup35p was shown to be able to be incorporated into existing aggregations even when three of the five oligopeptide repeats normally present were deleted. [25]
Function
It has been proposed that neurodegeneration caused by prions may be related to abnormal function of PrP. However, the physiological function of the prion protein remains a controversial matter. While data from in vitro experiments suggest many dissimilar roles, studies on PrP knockout mice have provided only limited information because these animals exhibit only minor abnormalities. In recent research done in mice, it was found that the cleavage of prions in peripheral nerves causes the activation of myelin repair in Schwann Cells. And that the lack of prions caused demyelination in those cells.[26]
PrP and long-term memory
There is evidence that PrP may have a normal function in maintenance of long-term memory.[27] Maglio and colleagues have shown that mice without the genes for normal cellular PrP protein have altered hippocampal long-term potentiation.[28]
PrP and stem cell renewal
A 2006 article from the Whitehead Institute for Biomedical Research indicates that PrP expression on stem cells is necessary for an organism's self-renewal of bone marrow. The study showed that all long-term hematopoietic stem cells expressed PrP on their cell membrane and that hematopoietic tissues with PrP-null stem cells exhibited increased sensitivity to cell depletion.[29]
Prion disease
| Affected animal(s) | Disease |
|---|---|
| sheep, goat | Scrapie[30] |
| cattle | Bovine spongiform encephalopathy (BSE), mad cow disease[30] |
| mink[30] | Transmissible mink encephalopathy (TME) |
| white-tailed deer, elk, mule deer, moose[30] | Chronic wasting disease (CWD) |
| cat[30] | Feline spongiform encephalopathy (FSE) |
| nyala, oryx, greater kudu[30] | Exotic ungulate encephalopathy (EUE) |
| ostrich[31] | Spongiform encephalopathy (Not been shown to be transmissible.) |
| human | Creutzfeldt–Jakob disease (CJD)[30] |
| iatrogenic Creutzfeldt-Jakob disease (iCJD) | |
| variant Creutzfeldt-Jakob disease (vCJD) | |
| familial Creutzfeldt-Jakob disease (fCJD) | |
| sporadic Creutzfeldt-Jakob disease (sCJD) | |
| Gerstmann–Sträussler–Scheinker syndrome (GSS)[30] | |
| Fatal familial insomnia (sFI)[32] | |
| Kuru[30] |
Prions cause neurodegenerative disease by aggregating extracellularly within the central nervous system to form plaques known as amyloid, which disrupt the normal tissue structure. This disruption is characterized by "holes" in the tissue with resultant spongy architecture due to the vacuole formation in the neurons.[33] Other histological changes include astrogliosis and the absence of an inflammatory reaction.[34] While the incubation period for prion diseases is generally quite long, once symptoms appear the disease progresses rapidly, leading to brain damage and death.[35] Neurodegenerative symptoms can include convulsions, dementia, ataxia (balance and coordination dysfunction), and behavioural or personality changes.
All known prion diseases, collectively called transmissible spongiform encephalopathies (TSEs), are untreatable and fatal.[36] A vaccine has been developed in mice, however, that may provide insight into providing a vaccine in humans to resist prion infections.[37] Additionally, in 2006 scientists announced that they had genetically engineered cattle lacking a necessary gene for prion production – thus theoretically making them immune to BSE,[38] building on research indicating that mice lacking normally occurring prion protein are resistant to infection by scrapie prion protein.[39]
Many different mammalian species can be affected by prion diseases, as the prion protein (PrP) is very similar in all mammals.[40] Due to small differences in PrP between different species it is unusual for a prion disease to be transmitted from one species to another. The human prion disease variant Creutzfeldt-Jakob disease, however, is believed to be caused by a prion which typically infects cattle, causing Bovine spongiform encephalopathy and is transmitted through infected meat.[41]
Transmission
It has been recognized that prion diseases can arise in three different ways: acquired, familial, or sporadic.[42] It is often assumed that the diseased form directly interacts with the normal form to make it rearrange its structure. One idea, the "Protein X" hypothesis, is that an as-yet unidentified cellular protein (Protein X) enables the conversion of PrPC to PrPSc by bringing a molecule of each of the two together into a complex.[43]
Current research suggests that the primary method of infection in animals is through ingestion. It is thought that prions may be deposited in the environment through the remains of dead animals and via urine, saliva, and other body fluids. They may then linger in the soil by binding to clay and other minerals.[44]
Sterilization
Infectious particles possessing nucleic acid are dependent upon it to direct their continued replication. Prions, however, are infectious by their effect on normal versions of the protein. Sterilizing prions therefore involves the denaturation of the protein to a state where the molecule is no longer able to induce the abnormal folding of normal proteins. Prions are generally quite resistant to proteases, heat, radiation, and formalin treatments,[45] although their infectivity can be reduced by such treatments. Effective prion decontamination relies upon protein hydrolysis or reduction or destruction of protein tertiary structure. Examples include bleach, caustic soda, and strong acidic detergents such as LpH.[46] 134°C (274°F) for 18 minutes in a pressurized steam autoclave may not be enough to deactivate the agent of disease.[47][48] Ozone sterilization is currently being studied as a potential method for prion denature and deactivation.[49] Renaturation of a completely denatured prion to infectious status has not yet been achieved, however partially denatured prions can be renatured to an infective status under certain artificial conditions.[50]
The World Health Organization recommends any of the following three procedures for the sterilization of all heat-resistant surgical instruments to ensure that they are not contaminated with prions:
- Immerse in a pan containing 1N NaOH and heat in a gravity-displacement autoclave at 121°C for 30 minutes; clean; rinse in water; and then perform routine sterilization processes.
- Immerse in 1N NaOH or sodium hypochlorite (20,000 parts per million available chlorine) for 1 hour; transfer instruments to water; heat in a gravity-displacement autoclave at 121°C for 1 hour; clean; and then perform routine sterilization processes.
- Immerse in 1N NaOH or sodium hypochlorite (20,000 parts per million available chlorine) for 1 hour; remove and rinse in water, then transfer to an open pan and heat in a gravity-displacement (121°C) or in a porous-load (134°C) autoclave for 1 hour; clean; and then perform routine sterilization processes.[51]
Debate
Whether prions are the agent which causes disease or merely a symptom caused by a different agent is still debated by a minority of researchers. The following sections describe several alternative hypotheses: some pertain to the composition of the infectious agent (protein-only, protein with other components, virus, or other), while others pertain to its mechanism of reproduction.
Protein hypothesis
Prior to the discovery of prions, it was thought that all pathogens used nucleic acids to direct their replication. The "protein hypothesis" states that a protein structure can replicate without the use of nucleic acid. This was initially controversial as it contradicts the so-called "central dogma of molecular biology", which describes nucleic acid as the central form of replicative information.
Evidence in favor of a protein hypothesis includes:[52]
- No virus particles, bacteria, or fungi have been conclusively associated with prion diseases, although Saccharomyces cerevisiae has been known to be associated with infectious, yet non-lethal prions, such as Sup35p.
- No nucleic acid has been conclusively associated with infectivity; agent is resistant to ultraviolet radiation
- No immune response to infection
- PrPSc experimentally transmitted between one species and another results in PrPSc with the amino-acid sequence of the recipient species, suggesting that replication of the donor agent does not occur
- Familial prion disease occurs in families with a mutation in the PrP gene, and mice with PrP mutations develop prion disease despite controlled conditions where transmission is prevented
- Animals lacking PrPC do not contract prion disease.
- Infectious prions can be formed de novo from purified non-infectious components, in the absence of gene-coding nucleic acids.[11]
Genetic factors
A gene for the normal protein has been identified: the PRNP gene.[53] In all inherited cases of prion disease, there is a mutation in the PRNP gene. Many different PRNP mutations have been identified and it is thought that the mutations somehow make PrPC more likely to change spontaneously into the abnormal PrPSc form.[54] Although this discovery puts a hole in the general prion hypothesis, that prions can only aggregate proteins of identical amino acid make up. These mutations can occur throughout the gene. Some mutations involve expansion of the octapeptide repeat region at the N-terminal of PrP. Other mutations that have been identified as a cause of inherited prion disease occur at positions 102, 117 & 198 (GSS), 178, 200, 210 & 232 (CJD) and 178 (Fatal Familial Insomnia, FFI). The cause of prion disease can be sporadic, genetic, and infectious, or a combination of these factors.[55] For example, in order to have scrapie, both an infectious agent and a susceptible genotype need to be present.[54]
Multi-component hypothesis
In 2007, biochemist Surachai Supattapone and his colleagues at Dartmouth College produced purified infectious prions de novo from defined components (PrPC, co-purified lipids, and a synthetic polyanionic molecule).[11] These researchers also showed that the polyanionic molecule required for prion formation was selectively incorporated into high-affinity complexes with PrP molecules, leading them to hypothesize that infectious prions may be composed of multiple host components, including PrP, lipid, and polyanionic molecules, rather than PrPSc alone.[56]
In 2010, Jiyan Ma and colleagues at The Ohio State University produced infectious prions from a recipe of bacterially expressed recombinant PrP, POPG phospholipid, and RNA, further supporting the multi-component hypothesis.[57] This finding is in contrast to studies that found minimal infectious prions produced from recombinant PrP alone.[58][59]
Heavy metal poisoning hypothesis
Recent reports suggest that imbalance of brain metal homeostasis is a significant cause of PrPSc-associated neurotoxicity, though the underlying mechanisms are difficult to explain based on existing information. Proposed hypotheses include a functional role for PrPC in metal metabolism, and loss of this function due to aggregation to the disease associated PrPSc form as the cause of brain metal imbalance. Other views suggest gain of toxic function by PrPSc due to sequestration of PrPC-associated metals within the aggregates, resulting in the generation of redox-active PrPSc complexes. The physiological implications of some PrPC-metal interactions are known, while others are still unclear. The pathological implications of PrPC-metal interaction include metal-induced oxidative damage, and in some instances conversion of PrPC to a PrPSc-like form.[60]
Viral hypothesis
The protein-only hypothesis has been criticised by those who feel that the simplest explanation of the evidence to date is viral.[61] For more than a decade, Yale University neuropathologist Laura Manuelidis has been proposing that prion diseases are caused instead by an unidentified "slow" virus. In January 2007, she and her colleagues published an article reporting to have found a virus in 10%, or less, of their scrapie-infected cells in culture.[62][63]
The virion hypothesis states that TSEs are caused by a replicable informational molecule (which is likely to be a nucleic acid) bound to PrP. Many TSEs, including scrapie and BSE, show strains with specific and distinct biological properties, a feature which supporters of the virion hypothesis feel is not explained by prions.
Evidence in favor of a viral hypothesis includes:[52]
- Strain variation: differences in prion infectivity, incubation, symptomology and progression among species resembles that seen between viruses, especially RNA viruses
- The long incubation and rapid onset of symptoms resembles lentiviruses, such as HIV-induced AIDS
- Viral-like particles that do not appear to be composed of PrP have been found in some of the cells of scrapie- or CJD-infected cell lines.[63]
Recent studies propagating TSE infectivity in cell-free reactions[64] and in purified component chemical reactions [11] strongly suggest against TSE viral nature. More recently, using a similar defined recipe of multiple components (PrP, POPG lipid, RNA), Jiyan Ma and colleagues generated infectious prions from recombinant PrP expressed from E. coli[57], casting further doubt on the viral hypothesis.
Fungi
Fungal prion proteins were discovered in the yeast Saccharomyces cerevisiae by Reed Wickner in the early 1990s. Subsequently, a prion has also been found in the fungus Podospora anserina. These prions behave similarly to PrP, but are generally non-toxic to their hosts. Susan Lindquist's group at the Whitehead Institute has argued that some of the fungal prions are not associated with any disease state, but may have a useful role; however, researchers at the NIH have also provided strong arguments demonstrating that fungal prions should be considered a diseased state.[65] Thus, the issue of whether fungal proteins are diseases, or have evolved for some specific functions still remains unresolved.[66]
As of 2010, there are 8 known prion proteins in fungi, 7 in Saccharomyces cerevisiae (Sup35, Rnq1, Ure2, Swi1, Mca1, Mot3, Cyc8) and one in Podospora anserina (HET-s).
Research into fungal prions has given strong support to the protein-only hypothesis for mammalian prions, since it has been demonstrated that purified protein extracted from cells with a prion state can convert the normal form of the protein into a misfolded form in vitro, and in the process, preserve the information corresponding to different strains of the prion state. It has also shed some light on prion domains, which are regions in a protein that promote the conversion into a prion. Fungal prions have helped to suggest mechanisms of conversion that may apply to all prions, though mammalian prions may operate by an independent mechanism.
| Fungal Prions | |||||
|---|---|---|---|---|---|
| Protein | Natural Host | Normal Function | Prion State | Prion Phenotype | Year Identified |
| Ure2p | Saccharomyces cerevisiae | Nitrogen catabolite repressor | [URE3] | Growth on poor nitrogen sources | 1994 |
| Sup35p | Saccharomyces cerevisiae | Translation termination factor | [PSI+] | Increased levels of nonsense suppression | 1994 |
| HET-S | Podospora anserina | Regulates heterokaryon incompatibility | [Het-s] | Heterokaryon formation between incompatible strains | |
| Rnq1p | Saccharomyces cerevisiae | Protein template factor | [RNQ+],[PIN+] | Promotes aggregation of other prions | |
| Mca1 | Saccharomyces cerevisiae | Putative Yeast Caspase | [MCA+] | Unknown | 2008 |
| Swi1 | Saccharomyces cerevisiae | chromatin remodeling | [SWI+] | poor growth on some carbon sources | 2008 |
| Cyc8 | Saccharomyces cerevisiae | transcriptional repressor | [OCT+] | transcriptional derepression of multiple genes | 2009 |
| Mot3 | Saccharomyces cerevisiae | Nuclear transcription factor | [MOT3+] | transcriptional derepression of anaerobic genes | 2009 |
- A putative prion protein, forming the [ISP+] element remains to be identified.
Potential treatments
Advancements in computer modeling have allowed for scientists to identify compounds which can serve as a treatment for prion caused diseases, such as one compound found to bind a cavity in the PrPC and stabilize the conformation, reducing the amount of harmful PrPSc.[67]
Recently, anti-prion antibodies capable of crossing the blood-brain-barrier and targetting cytosolic prion protein (an otherwise major obstacle in prion therapeutics) have been described [68]
See also
- Protein folding
- Protein Misfolding Cyclic Amplification
- Proteopathy
- Tertiary structure
Further reading
- Deadly Feasts: The "Prion" Controversy and the Public's Health[69], by Richard Rhodes
- The Pathological Protein: Mad Cow, Chronic Wasting, and Other Deadly Prion Diseases, Phillip Yam, 2003, Springer, ISBN 0-387-95508-9
- The Family That Couldn't Sleep by D. T. Max provides a history of prion diseases.
- The Prion Protein a special issue of the open-access journal Current Issues in Molecular Biology
- The Prion's Elusive Reason for Being
References
- ↑ "Prion". Oxford English Dictionary. Oxford University Press. 2nd ed. 1989.
- ↑ Ryan KJ, Ray CG, et al, ed (2004). Sherris Medical Microbiology (4th ed.). McGraw Hill. pp. 624–8. ISBN 0-8385-8529-9.
- ↑ "Stanley B. Prusiner - Autobiography". NobelPrize.org. http://nobelprize.org/nobel_prizes/medicine/laureates/1997/prusiner-autobio.html. Retrieved 2007-01-02.
- ↑ Prusiner SB (November 1998). "Prions". Proceedings of the National Academy of Sciences of the United States of America 95 (23): 13363–83. doi:10.1073/pnas.95.23.13363. PMID 9811807. PMC 33918. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=9811807. Retrieved 2010-02-28.
- ↑ Aguzzi A (January 2008). "Unraveling prion strains with cell biology and organic chemistry". Proceedings of the National Academy of Sciences of the United States of America 105 (1): 11–2. doi:10.1073/pnas.0710824105. PMID 18172195. PMC 2224168. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=18172195. Retrieved 2010-02-28.
- ↑ Masel J, Jansen VAA, Nowak MA (March 1999). "Quantifying the kinetic parameters of prion replication". Biophysical Chemistry 77 (2-3): 139-152. PMID 10326247.
- ↑ Dobson CM (February 2001). "The structural basis of protein folding and its links with human disease". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 356 (1406): 133–45. doi:10.1098/rstb.2000.0758. PMID 11260793. PMC 1088418. http://rstb.royalsocietypublishing.org/cgi/pmidlookup?view=long&pmid=11260793. Retrieved 2010-02-28.
- ↑ Li J, Browning S, Mahal SP, Oelschlegel AM, Weissmann C (February 2010). "Darwinian evolution of prions in cell culture". Science (New York, N.Y.) 327 (5967): 869–72. doi:10.1126/science.1183218. PMID 20044542. PMC 2848070. http://www.sciencemag.org/cgi/pmidlookup?view=long&pmid=20044542. Retrieved 2010-02-28. Lay summary.}}
- ↑ 9.0 9.1 9.2 Krull, Ira S.; Brian K. Nunnally (2004). Prions and mad cow disease. New York, N.Y: Marcel Dekker. pp. 6. ISBN 0-8247-4083-1. http://books.google.com/?id=WjeuaHopV5UC&pg=PA6&dq=Prions+and+mad+cow+disease+sedimentable&cd=1#v=onepage&q=. Retrieved 2010-02-28.
- ↑ Laurén J, Gimbel DA, Nygaard HB, Gilbert JW, Strittmatter SM (February 2009). "Cellular prion protein mediates impairment of synaptic plasticity by amyloid-beta oligomers". Nature 457 (7233): 1128–32. doi:10.1038/nature07761. PMID 19242475.
- ↑ 11.0 11.1 11.2 11.3 Deleault NR, Harris BT, Rees JR, Supattapone S (June 2007). "Formation of native prions from minimal components in vitro". Proceedings of the National Academy of Sciences of the United States of America 104 (23): 9741–6. doi:10.1073/pnas.0702662104. PMID 17535913. PMC 1887554. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=17535913. Retrieved 2010-02-28.
- ↑ Lindquist S, Krobitsch S, Li L, Sondheimer N (February 2001). "Investigating protein conformation-based inheritance and disease in yeast". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 356 (1406): 169–76. doi:10.1098/rstb.2000.0762. PMID 11260797. PMC 1088422. http://rstb.royalsocietypublishing.org/cgi/pmidlookup?view=long&pmid=11260797. Retrieved 2010-02-28.
- ↑ Alper T, Cramp WA, Haig DA, Clarke MC (May 1967). "Does the agent of scrapie replicate without nucleic acid?". Nature 214 (5090): 764–6. doi:10.1038/214764a0. PMID 4963878.
- ↑ Griffith JS (September 1967). "Self-replication and scrapie". Nature 215 (5105): 1043–4. doi:10.1038/2151043a0. PMID 4964084.
- ↑ Crick F (August 1970). "Central dogma of molecular biology". Nature 227 (5258): 561–3. doi:10.1038/227561a0. PMID 4913914.
- ↑ Taubes, Gary (December 1986). "The game of name is fame. But is it science?". Discover 7 (12): 28–41.
- ↑ "The Nobel Prize in Physiology or Medicine, 1997". NobelPrize.org. http://nobelprize.org/nobel_prizes/medicine/laureates/1997/. Retrieved 2010-02-28.
- ↑ Priola SA, Chesebro B, Caughey B (May 2003). "Biomedicine. A view from the top--prion diseases from 10,000 feet". Science (New York, N.Y.) 300 (5621): 917–9. doi:10.1126/science.1085920. PMID 12738843. http://www.sciencemag.org/cgi/pmidlookup?view=long&pmid=12738843. Retrieved 2010-02-28.
- ↑ Hegde RS, Mastrianni JA, Scott MR, et al. (February 1998). "A transmembrane form of the prion protein in neurodegenerative disease". Science 279 (5352): 827–34. doi:10.1126/science.279.5352.827. PMID 9452375. http://www.sciencemag.org/cgi/pmidlookup?view=long&pmid=9452375. Retrieved 2010-02-28.
- ↑ Brown DR, Qin K, Herms JW, et al. (1997). "The cellular prion protein binds copper in vivo". Nature 390 (6661): 684–7. doi:10.1038/37783. PMID 9414160.
- ↑ Weissmann C (November 2004). "The state of the prion". Nature Reviews. Microbiology 2 (11): 861–71. doi:10.1038/nrmicro1025. PMID 15494743.
- ↑ Málaga-Trillo E, Solis GP, Schrock Y, et al. (March 2009). "Regulation of embryonic cell adhesion by the prion protein". Plos Biology 7 (3): e55. doi:10.1371/journal.pbio.1000055. PMID 19278297. PMC 2653553. http://dx.plos.org/10.1371/journal.pbio.1000055. Retrieved 2010-02-28.
- ↑ Pan KM, Baldwin M, Nguyen J, et al. (December 1993). "Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins". Proceedings of the National Academy of Sciences of the United States of America 90 (23): 10962–6. doi:10.1073/pnas.90.23.10962. PMID 7902575. PMC 47901. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC47901/?tool=pubmed. Retrieved 2010-02-28.
- ↑ Baker, Harry F., and Rosalind M. Ridley, eds. Prion diseases. Totowa, N.J: Humana, 1996
- ↑ http://www.nature.com/emboj/journal/v20/n9/full/7593711a.html
- ↑ Abbott A (2010-01-24). "Healthy prions protect nerves". Nature. doi:10.1038/news.2010.29. http://www.nature.com/news/2010/100124/full/news.2010.29.html. Retrieved 2010-02-28.
- ↑ Shorter J, Lindquist S (June 2005). "Prions as adaptive conduits of memory and inheritance". Nature Reviews. Genetics 6 (6): 435–50. doi:10.1038/nrg1616. PMID 15931169.
- ↑ Maglio LE, Perez MF, Martins VR, Brentani RR, Ramirez OA (November 2004). "Hippocampal synaptic plasticity in mice devoid of cellular prion protein". Brain Research. Molecular Brain Research 131 (1-2): 58–64. doi:10.1016/j.molbrainres.2004.08.004. PMID 15530652. http://linkinghub.elsevier.com/retrieve/pii/S0169-328X(04)00399-7. Retrieved 2010-02-28.
- ↑ Zhang CC, Steele AD, Lindquist S, Lodish HF (February 2006). "Prion protein is expressed on long-term repopulating hematopoietic stem cells and is important for their self-renewal". Proceedings of the National Academy of Sciences of the United States of America 103 (7): 2184–9. doi:10.1073/pnas.0510577103. PMID 16467153. PMC 1413720. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=16467153. Retrieved 2010-02-28.
- ↑ 30.0 30.1 30.2 30.3 30.4 30.5 30.6 30.7 30.8 "90. Prions". ICTVdB Index of Viruses. U.S. National Institutes of Health website. 2002-02-14. http://www.ncbi.nlm.nih.gov/ICTVdb/Ictv/fs_prion.htm. Retrieved 2010-02-28.
- ↑ Hussein MF, Al-Mufarrej SI (2004). "Prion Diseases: A Review; II. Prion Diseases in Man and Animals." (pdf). Scientific Journal of King Faisal University (Basic and Applied Sciences) 5 (2): 139. http://www3.kfu.edu.sa/sjournal/eng/pdffiles/b526.pdf. Retrieved 2010-02-28.
- ↑ "BSE proteins may cause fatal insomnia.". BBC News. 1999-05-28. http://news.bbc.co.uk/2/hi/health/355297.stm. Retrieved 2010-02-28.
- ↑ Robbins SL, Cotran RS, Kumar V, Collins T, ed (1999). Robbins pathologic basis of disease. Philadelphia: Saunders. ISBN 0-7216-7335-X.
- ↑ Belay ED (1999). "Transmissible spongiform encephalopathies in humans". Annual Review of Microbiology 53: 283–314. doi:10.1146/annurev.micro.53.1.283. PMID 10547693. http://arjournals.annualreviews.org/doi/abs/10.1146/annurev.micro.53.1.283?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%3dncbi.nlm.nih.gov. Retrieved 2010-02-28.
- ↑ "Prion Diseases". US Centers for Disease Control. 2006-01-26. http://www.cdc.gov/ncidod/dvrd/prions/. Retrieved 2010-02-28.
- ↑ Gilch S, Winklhofer KF, Groschup MH, et al. (August 2001). "Intracellular re-routing of prion protein prevents propagation of PrP(Sc) and delays onset of prion disease". The EMBO Journal 20 (15): 3957–66. doi:10.1093/emboj/20.15.3957. PMID 11483499.
- ↑ New York University Medical Center and School of Medicine (2005-05-14). "Active Vaccine Prevents Mice From Developing Prion Disease". Science Daily. http://www.sciencedaily.com/releases/2005/05/050514111648.htm. Retrieved 2010-02-28.
- ↑ Weiss, Rick (2007-01-01). "Scientists Announce Mad Cow Breakthrough.". The Washington Post. http://www.washingtonpost.com/wp-dyn/content/article/2006/12/31/AR2006123100672.html. Retrieved 2010-02-28. "Scientists said yesterday that they have used genetic engineering techniques to produce the first cattle that may be biologically incapable of getting mad cow disease."
- ↑ Büeler H, Aguzzi A, Sailer A, et al. (July 1993). "Mice devoid of PrP are resistant to scrapie". Cell 73 (7): 1339–47. doi:10.1016/0092-8674(93)90360-3. PMID 8100741. http://linkinghub.elsevier.com/retrieve/pii/0092-8674(93)90360-3. Retrieved 2010-02-28.
- ↑ Collinge J (2001). "Prion diseases of humans and animals: their causes and molecular basis". Annual Review of Neuroscience 24: 519–50. doi:10.1146/annurev.neuro.24.1.519. PMID 11283320. http://arjournals.annualreviews.org/doi/abs/10.1146/annurev.neuro.24.1.519?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%3dncbi.nlm.nih.gov. Retrieved 2010-02-28.
- ↑ Ironside JW (March 2006). "Variant Creutzfeldt-Jakob disease: risk of transmission by blood transfusion and blood therapies". Haemophilia : the Official Journal of the World Federation of Hemophilia 12 Suppl 1: 8–15; discussion 26–8. doi:10.1111/j.1365-2516.2006.01195.x. PMID 16445812. http://www3.interscience.wiley.com/resolve/openurl?genre=article&sid=nlm:pubmed&issn=1351-8216&date=2006&volume=12&issue=&spage=8. Retrieved 2010-02-28.
- ↑ Groschup MH, Kretzschmar HA, eds. (2001). "Prion Diseases Diagnosis and Pathogeneis". Archives of Virology (New York: Springer) Suppl 16.
- ↑ Telling GC, Scott M, Mastrianni J, et al. (October 1995). "Prion propagation in mice expressing human and chimeric PrP transgenes implicates the interaction of cellular PrP with another protein". Cell 83 (1): 79–90. doi:10.1016/0092-8674(95)90236-8. PMID 7553876. http://linkinghub.elsevier.com/retrieve/pii/0092-8674(95)90236-8. Retrieved 2010-02-28.
- ↑ Johnson CJ, Pedersen JA, Chappell RJ, McKenzie D, Aiken JM (July 2007). "Oral transmissibility of prion disease is enhanced by binding to soil particles". PLoS Pathogens 3 (7): e93. doi:10.1371/journal.ppat.0030093. PMID 17616973. PMC 1904474. http://dx.plos.org/10.1371/journal.ppat.0030093. Retrieved 2010-02-28.
- ↑ Qin K, O'Donnell M, Zhao RY (August 2006). "Doppel: more rival than double to prion". Neuroscience 141 (1): 1–8. doi:10.1016/j.neuroscience.2006.04.057. PMID 16781817. http://linkinghub.elsevier.com/retrieve/pii/S0306-4522(06)00510-0. Retrieved 2010-02-28.
- ↑ Race RE, Raymond GJ (February 2004). "Inactivation of transmissible spongiform encephalopathy (prion) agents by environ LpH". Journal of Virology 78 (4): 2164–5. doi:10.1128/JVI.78.4.2164-2165.2004. PMID 14747583. PMC 369477. http://jvi.asm.org/cgi/pmidlookup?view=long&pmid=14747583. Retrieved 2010-02-28.
- ↑ Collins SJ, Lawson VA, Masters CL (2004). "Transmissible spongiform encephalopathies". Lancet 363 (9402): 51–61. doi:10.1016/S0140-6736(03)15171-9. PMID 14723996.
- ↑ Brown P, Rau EH, Johnson BK, Bacote AE, Gibbs CJ, Gajdusek DC (March 2000). "New studies on the heat resistance of hamster-adapted scrapie agent: threshold survival after ashing at 600 degrees C suggests an inorganic template of replication". Proceedings of the National Academy of Sciences of the United States of America 97 (7): 3418–21. doi:10.1073/pnas.050566797. PMID 10716712. PMC 16254. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=10716712. Retrieved 2010-02-28.
- ↑ "Ozone Sterilization". UK Health Protection Agency. 2005-04-14. http://web.archive.org/web/20080522065033/http://www.hpa.org.uk/hpa/news/articles/press_releases/2005/050414_ozone_sterilizer.htm. Retrieved 2010-02-28.
- ↑ Weissmann C, Enari M, Klöhn PC, Rossi D, Flechsig E (December 2002). "Transmission of prions". Proceedings of the National Academy of Sciences of the United States of America 99 Suppl 4: 16378–83. doi:10.1073/pnas.172403799. PMID 12181490. PMC 139897. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=12181490. Retrieved 2010-02-28.
- ↑ Sutton JM, Dickinson J, Walker JT, Raven ND (September 2006). "Methods to minimize the risks of Creutzfeldt-Jakob disease transmission by surgical procedures: where to set the standard?". Clinical Infectious Diseases : an Official Publication of the Infectious Diseases Society of America 43 (6): 757–64. doi:10.1086/507030. PMID 16912952. http://www.journals.uchicago.edu/doi/abs/10.1086/507030?url_ver=Z39.88-2003&rfr_id=ori:rid:crossref.org&rfr_dat=cr_pub%3dncbi.nlm.nih.gov. Retrieved 2010-02-28.
- ↑ 52.0 52.1 Baker & Ridley (1996). Prion Disease. New Jersey: Humana Press. ISBN 0-89603-342-2.
- ↑ Oesch B, Westaway D, Wälchli M, et al. (April 1985). "A cellular gene encodes scrapie PrP 27-30 protein". Cell 40 (4): 735–46. doi:10.1016/0092-8674(85)90333-2. PMID 2859120. http://linkinghub.elsevier.com/retrieve/pii/0092-8674(85)90333-2. Retrieved 2010-02-28.
- ↑ 54.0 54.1 Goldmann W (2008). "PrP genetics in ruminant transmissible spongiform encephalopathies". Veterinary Research 39 (4): 30. doi:10.1051/vetres:2008010. PMID 18284908. http://publications.edpsciences.org/10.1051/vetres:2008010. Retrieved 2010-03-02.
- ↑ Geissen M, Krasemann S, Matschke J, Glatzel M (July 2007). "Understanding the natural variability of prion diseases". Vaccine 25 (30): 5631–6. doi:10.1016/j.vaccine.2007.02.041. PMID 17391814. http://linkinghub.elsevier.com/retrieve/pii/S0264-410X(07)00219-8. Retrieved 2010-03-02.
- ↑ Geoghegan JC, Valdes PA, Orem NR, et al. (December 2007). "Selective incorporation of polyanionic molecules into hamster prions". The Journal of Biological Chemistry 282 (50): 36341–53. doi:10.1074/jbc.M704447200. PMID 17940287. http://www.jbc.org/cgi/pmidlookup?view=long&pmid=17940287. Retrieved 2010-02-28.
- ↑ 57.0 57.1 Wang F, Wang X, Yuan CG, Ma J (February 2010). "Generating a prion with bacterially expressed recombinant prion protein". Science (New York, N.Y.) 327 (5969): 1132–5. doi:10.1126/science.1183748. PMID 20110469. PMC 2893558. http://www.sciencemag.org/cgi/pmidlookup?view=long&pmid=20110469. Retrieved 2010-02-28.
- ↑ Legname G, Baskakov IV, Nguyen HO, et al. (July 2004). "Synthetic mammalian prions". Science (New York, N.Y.) 305 (5684): 673–6. doi:10.1126/science.1100195. PMID 15286374. http://www.sciencemag.org/cgi/pmidlookup?view=long&pmid=15286374. Retrieved 2010-02-28.
- ↑ Makarava N, Kovacs GG, Bocharova O, et al. (February 2010). "Recombinant prion protein induces a new transmissible prion disease in wild-type animals". Acta Neuropathologica 119 (2): 177–87. doi:10.1007/s00401-009-0633-x. PMID 20052481.
- ↑ Singh N et al. (2010). "Prion Protein and Metal Interaction: Physiological and Pathological Implications". The Prion Protein. Savanna Press. ISBN 978-0-9543335-2-2.
- ↑ Manuelidis L (March 2007). "A 25 nm virion is the likely cause of transmissible spongiform encephalopathies". Journal of Cellular Biochemistry 100 (4): 897–915. doi:10.1002/jcb.21090. PMID 17044041.
- ↑ "Pathogenic Virus Found in Mad Cow Cells". Yale. 2007-02-02. http://opa.yale.edu/news/article.aspx?status=301&id=1659. Retrieved 2010-02-28.
- ↑ 63.0 63.1 Manuelidis L, Yu ZX, Barquero N, Banquero N, Mullins B (February 2007). "Cells infected with scrapie and Creutzfeldt-Jakob disease agents produce intracellular 25-nm virus-like particles". Proceedings of the National Academy of Sciences of the United States of America 104 (6): 1965–70. doi:10.1073/pnas.0610999104. PMID 17267596. PMC 1794316. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=17267596. Retrieved 2010-02-28.
- ↑ Castilla J, Saá P, Hetz C, Soto C (April 2005). "In vitro generation of infectious scrapie prions". Cell 121 (2): 195–206. doi:10.1016/j.cell.2005.02.011. PMID 15851027. http://linkinghub.elsevier.com/retrieve/pii/S0092-8674(05)00156-X. Retrieved 2010-02-28.
- ↑ Dong J, Bloom JD, Goncharov V, et al. (November 2007). "Probing the role of PrP repeats in conformational conversion and amyloid assembly of chimeric yeast prions". The Journal of Biological Chemistry 282 (47): 34204–12. doi:10.1074/jbc.M704952200. PMID 17893150. PMC 2262835. http://www.jbc.org/cgi/pmidlookup?view=long&pmid=17893150. Retrieved 2010-02-28.
- ↑ Halfmann R, Alberti S, Lindquist S (2010). "Prions, protein homeostasis, and phenotypic diversity". Trends in Cell Biology 20 (3): 125–33. doi:10.1016/j.tcb.2009.12.003. PMID 20071174.
- ↑ Kuwata K, Nishida N, Matsumoto T, et al. (July 2007). "Hot spots in prion protein for pathogenic conversion". Proceedings of the National Academy of Sciences of the United States of America 104 (29): 11921–6. doi:10.1073/pnas.0702671104. PMID 17616582. PMC 1924567. http://www.pnas.org/cgi/pmidlookup?view=long&pmid=17616582. Retrieved 2010-02-28.
- ↑ http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0009804
- ↑ Deadly Feasts: The "Prion" Controversy and the Public's Health, Richard Rhodes, 1998, Touchstone, ISBN 0-684-84425-7
External links
General
- CDC – USA Centers for Disease Control and Prevention – information on prion diseases
- World Health Organisation – WHO information on prion diseases
- Prion Animation (Flash required)
Reports and committees
- The UK BSE Inquiry – Report of the UK public inquiry into BSE and variant CJD
- UK Spongiform Encephalopathy Advisory Committee (SEAC)
Genetics
- Mammalian prion classification International Committee on Taxonomy of Viruses – ICTVdb
- Online Mendelian Inheritance in Man: Prion protein – PrP, inherited prion disease and transgenic animal models.
- The Surprising World of Prion Biology--A New Mechanism of Inheritance on-line lecture by Susan Lindquist
Research
- Institute for Neurodegenerative Diseases – labs studying prion diseases, run by Stanley B. Prusiner, MD
- Prion Disease Database (PDDB) - Comprehensive transcriptome resource for systems biology research in prion diseases.
Other
- UCSF Memory and Aging Center – medical center for diagnosis and care of people with prion disease and research into origin and treatment of prion diseases. (YouTube channel)
|
||||||||