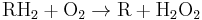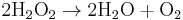Peroxisome

Peroxisomes are organelles present in almost all eukaryotic cells. They participate in the metabolism of fatty acids and many other metabolites. Peroxisomes harbour enzymes that rid the cell of toxic peroxides. Peroxisomes are bound by a single membrane that separates their contents from the cytosol (the internal fluid of the cell) and contain membrane proteins critical for various functions, such as importing proteins into the organelles and aiding in proliferation. Peroxisomes can replicate by enlarging and then dividing. Peroxisomes were identified as organelles by the Belgian cytologist Christian de Duve in 1967[1] after they had been first described in a PhD thesis of Rhodin a decade earlier.[2]
Contents |
Occurrence and evolution
Peroxisomes are found in virtually all eukaryotic cells. Peroxisomes contain enzymes for certain oxidative reactions, like the beta-oxidation of very-long-chain fatty acids. Prokaryotes lack peroxisomes. The enzymatic content of peroxisomes varies across species, but the presence of certain proteins common to many species has been used to suggest an endosymbiotic origin; that is, peroxisomes evolved from bacteria that invaded larger cells as parasites, and very gradually evolved a symbiotic relationship.[3] However, this view has been challenged by recent discoveries.[4] For example, peroxisome-less mutants can restore peroxisomes upon introduction of the wild-type gene, and peroxisomes have been observed to be formed from the endoplasmic reticulum.[5]
An evolutionary analysis of the peroxisomal proteome found homologies between the peroxisomal import machinery and the ERAD pathway in the endoplasmic reticulum [6], along with a number of metabolic enzymes that were likely recruited from the mitochondria.[7] These results indicate that the peroxisome does not have an endosymbiotic origin; instead, it likely originates from the ER, and its proteins were recruited from pools existing within the primitive eukaryote, as quoted in the science textbook Biozone. However, latest research are suggests that the peroxisome may have had an actinobacterial origin [8].
Other organelles of the Microbody family related to peroxisomes include glyoxysomes of plants and filamentous fungi, glycosomes of kinetoplastids [9] and Woronin bodies of filamentous fungi.
Function
Peroxisomes contain oxidative enzymes, such as catalase, D-amino acid oxidase, and uric acid oxidase.[10] However the last enzyme is absent in humans, explaining the disease known as gout, caused by the accumulation of uric acid. Certain enzymes within the peroxisome, by using molecular oxygen, remove hydrogen atoms from specific organic substrates (labeled as R), in an oxidative reaction, producing hydrogen peroxide (H2O2, itself toxic):
Catalase, another enzyme in the peroxisome, in turn uses this H2O2 to oxidize other substrates, including phenols, formic acid, formaldehyde, and alcohol, by means of the peroxidation reaction:
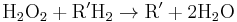 , thus eliminating the poisonous hydrogen peroxide in the process.
, thus eliminating the poisonous hydrogen peroxide in the process.
This reaction is important in liver and kidney cells, where the peroxisomes detoxify various toxic substances that enter the blood. About 25% of the ethanol we drink is oxidized to acetaldehyde in this way. In addition, when excess H2O2 accumulates in the cell, catalase converts it to H2O through this reaction:
A major function of the peroxisome is the breakdown of fatty acid molecules, in a process called beta-oxidation. In this process, the fatty acids are broken down two carbons at a time, converted to Acetyl-CoA, which is then transported back to the cytosol for further use. In animal cells, beta-oxidation can also occur in the mitochondria. In yeast and plant cells, this process is exclusive for the peroxisome.[11]
The first reactions in the formation of plasmalogen in animal cells also occur in peroxisomes. Plasmalogen is the most abundant phospholipid in myelin. Deficiency of plasmalogens causes profound abnormalities in the myelination of nerve cells, which is one of the reasons that many peroxisomal disorders lead to neurological disease (adrenoleukodystrophy).[11]
Peroxisomes also play a role in the production of bile acids and proteins.
In higher plants, peroxisomes contain also a complex battery of antioxidative enzymes such as superoxide dismutase, the components of the ascorbate-glutathione cycle, and the NADP-dehydrogenases of the pentose-phosphate pathway. It has been demonstrated the generation of superoxide (O2•-) and nitric oxide (•NO) radicals.[12][13]
The peroxisome of plant cells is polarised when fighting fungal penetration. Infection causes a glucosinolate molecule to play an antifungal role to be made and delivered to the outside of the cell through the action of the peroxisomal proteins (PEN2 and PEN3).[14]
Protein import
Proteins are selectively imported into peroxisomes. Since the organelles contain no DNA or ribosomes and, thus, have no means of producing proteins, all of their proteins must be imported across the membrane. It is believed that necessary proteins enter through the endoplasmic reticulum during biogenesis as well as through membrane proteins.
A specific protein signal (PTS or peroxisomal targeting signal) of three amino acids at the C-terminus of many peroxisomal proteins signals the membrane of the peroxisome to import them into the organelle. Other peroxisomal proteins contain a signal at the N-terminus. There are at least 32 known peroxisomal proteins, called peroxins,[15] which participate in the process of importing proteins by means of ATP hydrolysis. Proteins do not have to unfold to be imported into the peroxisome. The protein receptors, the peroxins PEX5 and PEX7, accompany their cargoes (containing a PTS1 or a PTS2, respectively) all the way into the peroxisome where they release the cargo and then return to the cytosol - a step named recycling. Overall, the import cycle is referred to as the extended shuttle mechanism. Evidence now indicates that ATP hydrolysis is required for the recycling of receptors to the cytosol. Also, ubiquitination appears to be crucial for the export of PEX5 from the peroxisome, to the cytosol. Little is known about the import of PEX7, although it has helper proteins that have been shown to be ubiquitinated. It is interesting to note that Pex7 helper proteins share many similarities with Pex5, and, in higher eukaryotes, they are functionally replaced by a long isoform of PEX5 (PEX5L).
Deficiencies
Peroxisomal disorders are a class of conditions that lead to disorders of lipid metabolism and diseases of the nervous system. Well-known examples are X-linked adrenoleukodystrophy, the most frequent, and Zellweger syndrome [16][17]. Peroxisomes matrix proteins are synthesized on free ribosomes in the cytosol and are imported post-translationally in pre-existing vesicles.
Genes
Genes that encode peroxisomal proteins include:
- PEX1
- PEX2 - PXMP3
- PEX3
- PEX5
- PEX6
- PEX7
- PEX10
- PEX11A, PEX11B, PEX11G
- PEX12
- PEX13
- PEX14
- PEX16
- PEX19
- PEX26
- PEX28
- PEX30
- PEX31
References
- ↑ de Duve C (1969). "The peroxisome: a new cytoplasmic organelle". Proc. R. Soc. Lond., B, Biol. Sci. 173 (30): 71–83. PMID 4389648.
- ↑ Rhodin, J (1954). "Correlation of ultrastructural organization and function in normal and experimentally changed proximal tubule cells of the mouse kidney". Doctorate Thesis. Karolinska Institutet, Stockholm.
- ↑ Lazarow PB, Fujiki Y (1985). "Biogenesis of peroxisomes". Annu. Rev. Cell Biol. 1: 489–530. doi:10.1146/annurev.cb.01.110185.002421. PMID 3916321.
- ↑ Fagarasanu A, Fagarasanu M, Rachubinski, RA (2007). "Maintaining peroxisome populations: a story of division and inheritance". Annu. Rev. Cell Dev. Biol. 23: 321–344. doi:10.1146/annurev.cellbio.23.090506.123456. PMID 17506702.
- ↑ Hoepfner D, Schildknegt D, Braakman I, Philippsen P, Tabak HF (2005). "Contribution of the endoplasmic reticulum to peroxisome formation". Cell 122 (1): 85–95. doi:10.1016/j.cell.2005.04.025. PMID 16009135.
- ↑ Schlüter A, Fourcade S, Ripp R, Mandel JL, Poch O, Pujol A (2006). "The evolutionary origin of peroxisomes: an ER-peroxisome connection". Mol Biol Evol 23(4) (4): 838–45. doi:10.1093/molbev/msj103. PMID 16452116.
- ↑ Gabaldón T, Snel B, van Zimmeren F, Hemrika W, Tabak H, Huynen MA (2006). "Origin and evolution of the peroxisomal proteome". Biol. Direct 1: 8. doi:10.1186/1745-6150-1-8. PMID 16556314.
- ↑ Duhita, et al; Le, HA; Satoshi, S; Kazuo, H; Daisuke, M; Takao, S (2009). "The origin of peroxisomes: The possibility of an actinobacterial symbiosis.". Gene 450 (1-2): 18–24. doi:10.1016/j.gene.2009.09.014. PMID 19818387.
- ↑ Blattner, J.; Swinkels, B; Dörsam, H; Prospero, T; Subramani, S; Clayton, C (1992). "Glycosome assembly in trypanosomes: variations in the acceptable degeneracy of a COOH-terminal microbody targeting signal". The Journal of Cell Biology 119 (5): 1129. doi:10.1083/jcb.119.5.1129. PMID 1447292
- ↑ del Río LA, Sandalio LM, Palma JM, Bueno P, Corpas FJ (1992). "Metabolism of oxygen radicals in peroxisomes and cellular implications". Free Radic. Biol. Med. 13 (5): 557–80. doi:10.1016/0891-5849(92)90150-F. PMID 1334030.
- ↑ 11.0 11.1 "Peroxisomes" (chapter 12), Bruce Alberts, Alexander Johnson, Julian Lewis, Martin Raff, Keith Roberts, and Peter Walter (2002), Molecular Biology of the Cell, Fourth Edition, ISBN 0-8153-3218-1
- ↑ Corpas F.J., Barroso, J.B., del Río, L.A. (2001). "Peroxisomes as a source of reactive oxygen species and nitric oxide signal molecules in plant cells". Trends Plant Sci 6 (4): 145–150. doi:10.1016/S1360-1385(01)01898-2. PMID 11286918.
- ↑ Corpas FJ et al. (2004). "Cellular and subcellular localization of endogenous nitric oxide in young and senescent pea plants". Plant Physiol 136 (1): 2722–2733. doi:10.1104/pp.104.042812. PMID 15347796.
- ↑ Bednarek P, Pislewska-Bednarek M, Svatos A, et al (December 2008). "A Glucosinolate Metabolism Pathway in Living Plant Cells Mediates Broad-Spectrum Antifungal Defense". Science 323 (5910): 101. doi:10.1126/science.1163732. PMID 19095900.
- ↑ Saleem RA, Smith JJ, Aitchison JD (2006). "Proteomics of the peroxisome". Biochim. Biophys. Acta 1763 (12): 1541–51. doi:10.1016/j.bbamcr.2006.09.005. PMID 17050007. PMC 1858641. http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=1858641&blobtype=pdf.
- ↑ Depreter M, Espeel M, Roels F (2003). Human peroxisomal disorders, Microsc Res Techn 61: 203-223
- ↑ Roels F, De Bie S, Schutgens RBH, Besley GTN (1995). Diagnosis of human peroxisomal disorders. A handbook. J Inh Metab Dis 18, suppl 1: 1-229
External links
![]() This article incorporates public domain material from the NCBI document "Science Primer".
This article incorporates public domain material from the NCBI document "Science Primer".
|
||||||||||||||||||||||||||