Microscopy
Microscopy is the technical field of using microscopes to view samples or objects that cannot be seen with the unaided eye(objects that are not within the resolution range of the normal eye). There are three well-known branches of microscopy, optical, electron, and scanning probe microscopy.
Optical and electron microscopy involve the diffraction, reflection, or refraction of electromagnetic radiation/electron beams interacting with the subject of study, and the subsequent collection of this scattered radiation in order to build up an image. This process may be carried out by wide-field irradiation of the sample (for example standard light microscopy and transmission electron microscopy) or by scanning of a fine beam over the sample (for example confocal laser scanning microscopy and scanning electron microscopy). Scanning probe microscopy involves the interaction of a scanning probe with the surface or object of interest. The development of microscopy revolutionized biology and remains an essential tool in that science, along with many others including materials science and forensic engineering where microstructure and surface details such as trace evidence are important for understanding properties or the causes of accidents.

Optical microscopy

Optical or light microscopy involves passing visible light transmitted through or reflected from the sample through a single or multiple lenses to allow a magnified view of the sample.[1] The resulting image can be detected directly by the eye, imaged on a photographic plate or captured digitally. The single lens with its attachments, or the system of lenses and imaging equipment, along with the appropriate lighting equipment, sample stage and support, makes up the basic light microscope. The most recent development is the digital microscope, which uses a CCD camera to focus on the exhibit of interest. The image is shown on a computer screen since the camera is attached to it via a USB port, so eye-pieces are unnecessary.
Limitations
Limitations of standard optical microscopy (bright field microscopy) lie in three areas;
- The technique can only image dark or strongly refracting objects effectively.
- Diffraction limits resolution to approximately 0.2 micrometre (see: microscope).
- Out of focus light from points outside the focal plane reduces image clarity.
Live cells in particular generally lack sufficient contrast to be studied successfully, internal structures of the cell are colourless and transparent. The most common way to increase contrast is to stain the different structures with selective dyes, but this involves killing and fixing the sample. Staining may also introduce artifacts, apparent structural details that are caused by the processing of the specimen and are thus not a legitimate feature of the specimen.
These limitations have all been overcome to some extent by specific microscopy techniques that can non-invasively increase the contrast of the image. In general, these techniques make use of differences in the refractive index of cell structures. It is comparable to looking through a glass window: you (bright field microscopy) don't see the glass but merely the dirt on the glass. There is however a difference as glass is a denser material, and this creates a difference in phase of the light passing through. The human eye is not sensitive to this difference in phase but clever optical solutions have been thought out to change this difference in phase into a difference in amplitude (light intensity).
Techniques
In order to improve specimen contrast or highlight certain structures in a sample special techniques must be used. A huge selection of microscopy techniques are available to increase contrast or label a sample.
 Bright field illumination, sample contrast comes from absorbance of light in the sample. |
 Cross-polarized light illumination, sample contrast comes from rotation of polarized light through the sample. |
 Dark field illumination, sample contrast comes from light scattered by the sample. |
 Phase contrast illumination, sample contrast comes from interference of different path lengths of light through the sample. |
Bright field
Bright field microscopy is the simplest of all the light microscopy techniques. Sample illumination is via transmitted white light, i.e. illuminated from below and observed from above. Limitations include low contrast of most biological samples and low apparent resolution due to the blur of out of focus material. The simplicity of the technique and the minimal sample preparation required are significant advantages.
Oblique illumination
The use of oblique (from the side) illumination gives the image a 3-dimensional appearance and can highlight otherwise invisible features. A more recent technique based on this method is Hoffmann's modulation contrast, a system found on inverted microscopes for use in cell culture. Oblique illumination suffers from the same limitations as bright field microscopy (low contrast of many biological samples; low apparent resolution due to out of focus objects), but may highlight otherwise invisible structures.
Dark field
Dark field microscopy is a technique for improving the contrast of unstained, transparent specimens.[2] Dark field illumination uses a carefully aligned light source to minimize the quantity of directly-transmitted (unscattered) light entering the image plane, collecting only the light scattered by the sample. Darkfield can dramatically improve image contrast—especially of transparent objects – while requiring little equipment setup or sample preparation. However, the technique does suffer from low light intensity in final image of many biological samples, and continues to be affected by low apparent resolution.
Rheinberg illumination is a special variant of dark field illumination in which transparent, colored filters are inserted just before the condenser so that light rays at high aperture are differently colored than those at low aperture (i.e. the background to the specimen may be blue while the object appears self-luminous yellow). Other color combinations are possible but their effectiveness is quite variable.[3]
Dispersion staining
Dispersion staining is an optical technique that results in a colored image of a colorless object. This is an optical staining technique and requires no stains or dyes to produce a color effect. There are five different microscope configurations used in the broader technique of dispersion staining. They include brightfield Becke` line, oblique, darkfield, phase contrast, and objective stop dispersion staining.
Phase contrast
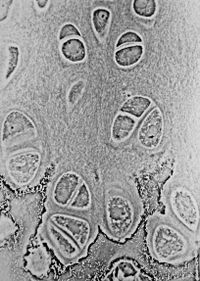
- In electron microscopy: Phase-contrast imaging
More sophisticated techniques will show proportional differences in optical density . Phase contrast is a widely used technique that shows differences in refractive index as difference in contrast. It was developed by the Dutch physicist Frits Zernike in the 1930s (for which he was awarded the Nobel Prize in 1953). The nucleus in a cell for example will show up darkly against the surrounding cytoplasm. Contrast is excellent; however it is not for use with thick objects. Frequently, a halo is formed even around small objects, which obscures detail. The system consists of a circular annulus in the condenser, which produces a cone of light. This cone is superimposed on a similar sized ring within the phase-objective. Every objective has a different size ring, so for every objective another condenser setting has to be chosen. The ring in the objective has special optical properties: it first of all reduces the direct light in intensity, but more importantly, it creates an artificial phase difference of about a quarter wavelength. As the physical properties of this direct light have changed, interference with the diffracted light occurs, resulting in the phase contrast image.
Differential interference contrast
Superior and much more expensive is the use of interference contrast. Differences in optical density will show up as differences in relief. A nucleus within a cell will actually show up as a globule in the most often used differential interference contrast system according to Georges Nomarski. However, it has to be kept in mind that this is an optical effect, and the relief does not necessarily resemble the true shape! Contrast is very good and the condenser aperture can be used fully open, thereby reducing the depth of field and maximizing resolution.
The system consists of a special prism (Nomarski prism, Wollaston prism) in the condenser that splits light in an ordinary and an extraordinary beam. The spatial difference between the two beams is minimal (less than the maximum resolution of the objective). After passage through the specimen, the beams are reunited by a similar prism in the objective.
In a homogeneous specimen, there is no difference between the two beams, and no contrast is being generated. However, near a refractive boundary (say a nucleus within the cytoplasm), the difference between the ordinary and the extraordinary beam will generate a relief in the image. Differential interference contrast requires a polarized light source to function; two polarizing filters have to be fitted in the light path, one below the condenser (the polarizer), and the other above the objective (the analyzer).
Note: In cases where the optical design of a microscope produces an appreciable lateral separation of the two beams we have the case of classical interference microscopy, which does not result in relief images, but can nevertheless be used for the quantitative determination of mass-thicknesses of microscopic objects.
Interference reflection microscopy
An additional technique using interference is interference reflection microscopy (also known as reflected interference contrast, or RIC). It is used to examine the adhesion of cells to a glass surface, using polarized light of a narrow range of wavelengths to be reflected whenever there is an interface between two substances with different refractive indices. Whenever a cell is attached to the glass surface, reflected light from the glass and from the attached cell will interfere, while if there is no cell attached to the glass, there will be no interference.
Interference reflection microscopy can be obtained by using the same elements used by DIC, but without the prisms. Also, the light that is being detected is reflected and not transmitted as it is when DIC is employed.
Fluorescence
When certain compounds are illuminated with high energy light, they then emit light of a different, lower frequency. This effect is known as fluorescence. Often specimens show their own characteristic autofluorescence image, based on their chemical makeup.
This method is of critical importance in the modern life sciences, as it can be extremely sensitive, allowing the detection of single molecules. Many different fluorescent dyes can be used to stain different structures or chemical compounds. One particularly powerful method is the combination of antibodies coupled to a fluorochrome as in immunostaining. Examples of commonly used fluorochromes are fluorescein or rhodamine. The antibodies can be made tailored specifically for a chemical compound. For example, one strategy often in use is the artificial production of proteins, based on the genetic code (DNA). These proteins can then be used to immunize rabbits, which then form antibodies which bind to the protein. The antibodies are then coupled chemically to a fluorochrome and then used to trace the proteins in the cells under study.
Highly-efficient fluorescent proteins such as the green fluorescent protein (GFP) have been developed using the molecular biology technique of gene fusion, a process that links the expression of the fluorescent compound to that of the target protein. Piston DW, Patterson GH, Lippincott-Schwartz J, Claxton NS, Davidson MW (2007). "Nikon MicroscopyU: Introduction to Fluorescent Proteins". Nikon MicroscopyU. http://www.microscopyu.com/articles/livecellimaging/fpintro.html. Retrieved 2007-08-22. This combined fluorescent protein is, in general, non-toxic to the organism and rarely interferes with the function of the protein under study. Genetically modified cells or organisms directly express the fluorescently-tagged proteins, which enables the study of the function of the original protein in vivo.
Since fluorescence emission differs in wavelength (color) from the excitation light, an ideal fluorescent image shows only the structure of interest that was labeled with the fluorescent dye. This high specificity led to the widespread use of fluorescence light microscopy in biomedical research. Different fluorescent dyes can be used to stain different biological structures, which can then be detected simultaneously, while still being specific due to the individual color of the dye.
To block the excitation light from reaching the observer or the detector, filter sets of high quality are needed. These typically consist of an excitation filter selecting the range of excitation wavelengths, a dichroic mirror, and an emission filter blocking the excitation light. Most fluorescence microscopes are operated in the Epi-illumination mode (illumination and detection from one side of the sample) to further decrease the amount of excitation light entering the detector.
See also total internal reflection fluorescence microscope.
Confocal
Confocal microscopy generates the image in a completely different way to normal "wide-field" microscopes. Using a scanning point of light instead of full sample illumination confocal microscopy gives slightly higher resolution, and significant improvements in optical sectioning by blocking the influence of out-of-focus light that would otherwise degrade the image. Confocal microscopy is, therefore, commonly used where 3D structure is important.
Deconvolution
Fluorescence microscopy is extremely powerful due to its ability to show specifically labeled structures within a complex environment and also because of its inherent ability to provide three-dimensional information of biological structures. However, this information is blurred by the fact that, upon illumination, all fluorescently labeled structures emit light no matter whether they are in focus or not. This means that an image of a certain structure is always blurred by the contribution of light from structures that are out of focus. This phenomenon becomes apparent as a loss of contrast especially when using objectives with a high resolving power, typically oil immersion objectives with a high numerical aperture.
However, this phenomenon is not caused by random processes such as light scattering but can be relatively well defined by the optical properties of the image formation in the microscope imaging system. If one considers a small fluorescent light source (essentially a bright spot), light coming from this spot spreads out the further out of focus one is. Under ideal conditions, this produces a sort of "hourglass" shape of this point source in the third (axial) dimension. This shape is called the point spread function (PSF) of the microscope imaging system. Since any fluorescence image is made up of a large number of such small fluorescent light sources, the image is said to be "convolved by the point spread function".
Knowing this point spread function[4] means that it is possible to reverse this process to a certain extent by computer-based methods commonly known as deconvolution microscopy.[5] There are various algorithms available for 2D or 3D deconvolution. They can be roughly classified in nonrestorative and restorative methods. While the nonrestorative methods can improve contrast by removing out-of-focus light from focal planes, only the restorative methods can actually reassign light to its proper place of origin. This can be an advantage over other types of 3D microscopy such as confocal microscopy, because light is not thrown away but reused. For 3D deconvolution, one typically provides a series of images derived from different focal planes (called a Z-stack) plus the knowledge of the PSF, which can be derived either experimentally or theoretically from knowing all contributing parameters of the microscope.
Sub-diffraction techniques
It is well known that there is a spatial limit to which light can focus: approximately half of the wavelength of the light one is using. But this is not a true barrier, because this diffraction limit is only true in the far-field and localization precision can be increased with many photons and careful analysis; and like the sound barrier, the diffraction barrier is breakable. This section explores some approaches to imaging objects smaller than ~250 nm. In 1978, first theoretical ideas have been developed to break this barrier by using a 4Pi microscope as a confocal laser scanning fluorescence microscope where the light is focused ideally from all sides to a common focus that is used to scan the object by 'point-by-point' excitation combined with 'point-by-point' detection [6]. Most of the following information was gathered (with permission) from a chemistry blog's review of sub-diffraction microscopy techniques Part I and Part II. For a review, see also reference [7].
Vertico SMI - SPDMphymod Superresolution Microscopy
Localization Microscopy/Spatially Structured Illumination
Around 1995, Christoph Cremer commenced with the development of a light microscopic process, which achieved a substantially improved size resolution of cellular nanostructures stained with a fluorescent marker. This time he employed the principle of wide field microscopy combined with structured laser illumination (spatially modulated illumination, SMI[8]. In addition, this technology is no longer subjected to the speed limitations of the focusing microscopy so that it becomes possible to undertake 3D analyses of whole cells within short observation times (at the moment around a few seconds).
Also since around 1995, Christoph Cremer developed and realized new fluorescence-based wide-field microscopy approaches that had as their goal the improvement of the effective optical resolution (in terms of the smallest detectable distance between two localized objects) down to a fraction of the conventional resolution (spectral precision distance/position determination microscopy, SPDM).
Combining SPDM and SMI, known as Vertico-SMI microscopy[9] Christoph Cremer can currently achieve a resolution of approx. 10 nm in 2D and 40 nm in 3D in wide field images of whole living cells[10]. The Vertico SMI is currently the fastest optical 3D nanoscope for the three-dimensional structural analysis of whole cells world-wide.
The Vertico SMI works with high recording speed and processes a complete 3D stack in 40-second (2000 frames), the very fast image processing based on specific proprietary algorithms makes the image available after 2min/3min (1-/2-color). The specific wide-field technique captures very large areas up to 5000 µm2.
|
Single YFP molecule detection in a human cancer cell. Typical distance measurements in the 15 nm range (5 nm standard deviation) |
Co-localisation microscopy (2CLM) with GFP and RFP fusion proteins (nucleus of a bone cancer cell) 120.000 localized molecules in a widefield area(470 µm2) |
Use of standard dyes like normal GFP
In 2008, Cremer´s lab discovered that super resolution microscopy was also possible for many standard fluorescent dyes like GFP, Alexa dyes and fluorescein molecules, provided certain photo-physical conditions are present. Using his specific localization microscopy called SPDMPhymod, it is possible to detect and count two different fluorescent molecule types (this technology is referred to as 2CLM, 2 Color Localization Microscopy)[11]
Near-field scanning
Near-field scanning is also called NSOM. Probably the most conceptual way to break the diffraction barrier is to use a light source and/or a detector that is itself nanometer in scale. Diffraction as we know it is truly a far-field effect: The light from an aperture is the Fourier transform of the aperture in the far-field.[12] But, in the near-field, all of this is not necessarily the case. Near-field scanning optical microscopy (NSOM) forces light through the tiny tip of a pulled fiber — and the aperture can be on the order of tens of nanometers.[13] When the tip is brought to nanometers away from a molecule, the resolution is limited not by diffraction but by the size of the tip aperture (because only that one molecule will see the light coming out of the tip). An image can be built by a raster scan of the tip over the surface to create an image.
The main down-side to NSOM is the limited number of photons you can force out a tiny tip, and the minuscule collection efficiency (if one is trying to collect fluorescence in the near-field). Other techniques such as ANSOM (see below) try to avoid this drawback.
Local enhancement / ANSOM / bowties
Instead of forcing photons down a tiny tip, some techniques create a local bright spot in an otherwise diffraction-limited spot. ANSOM is apertureless NSOM: it uses a tip very close to a fluorophore to enhance the local electric field the fluorophore sees.[14] Basically, the ANSOM tip is like a lightning rod which creates a hot spot of light.
Bowtie nanoantennas have been used to greatly and reproducibly enhance the electric field in the nanometer gap between the tips two gold triangles. Again, the point is to enhance a very small region of a diffraction-limited spot, thus improving the mismatch between light and nanoscale objects—and breaking the diffraction barrier.[15]
Stimulated emission depletion
Stefan Hell at the Max Planck Institute for Biophysical Chemistry - Göttingen (Germany) developed STED microscopy (stimulated emission depletion), which uses two laser pulses. The first pulse is a diffraction-limited spot that is tuned to the absorption wavelength, so excites any fluorophores in that region; an immediate second pulse is red-shifted to the emission wavelength and stimulates emission back to the ground state before, thereby depleting the excited state of any fluorophores in this depletion pulse. The trick is that the depletion pulse goes through a phase modulator that makes the pulse illuminate the sample in the shape of a donut, so the outer part of the diffraction limited spot is depleted and the small center can still fluoresce. By saturating the depletion pulse, the center of the donut gets smaller and smaller until they can get resolution of tens of nanometers.[16]
This technique also requires a raster scan like NSOM and standard confocal laser scanning microscopy.
Fitting the point-spread function
Fitting the point-spread function (also called PSF). The methods above (and below) use experimental techniques to circumvent the diffraction barrier, but one can also use crafty analysis to increase the ability to know where a nanoscale object is located. The image of a point source on a charge-coupled device camera is called a point-spread function (PSF), which is limited by diffraction to be no less than approximately half the wavelength of the light. But it is possible to simply fit that PSF with a Gaussian to locate the center of the PSF — and thus the location of the fluorophore. The precision by which this technique can locate the center depends on the number of photons collected (as well as the CCD pixel size and other factors).[17] This concept was first used to achieve resolution beyond the diffraction limit with single molecules by Van Oijen et al. in 1998 (Chem. Phys. Lett. V.292, p183). Subsequently at room temperature, groups like the Selvin lab and many others have employed this analysis to localize single fluorophores to a few nanometers. This, of course, requires careful measurements and collecting many photons.
PALM, STORM
What fitting a PSF is to localization, photo-activated localization microscopy (PALM) is to "resolution"—this term is here used loosely to mean measuring the distance between objects, not true optical resolution. Eric Betzig and colleagues developed PALM;[18] Xiaowei Zhuang at Harvard used a similar techniques and calls it STORM: stochastic optical reconstruction microscopy.[19] Sam Hess at University of Maine developed the technique simultaneously. The basic premise of both techniques is to fill the imaging area with many dark fluorophores that can be photoactivated into a fluorescing state by a flash of light. Because photoactivation is stochastic, only a few, well-separated molecules "turn on." Then Gaussians are fit to their PSFs to high precision (see section above). After the few bright dots photobleach, another flash of the photoactivating light activates random fluorophores again and the PSFs are fit of these different well-spaced objects. This process is repeated many times, building up an image molecule-by-molecule; and, because the molecules were localized at different times, the "resolution" of the final image can be much higher than that limited by diffraction.
The major problem with these techniques is that to get these beautiful pictures, it takes on the order of hours to collect the data. This is not the technique to study dynamics (fitting the PSF is better for that).
Structured illumination
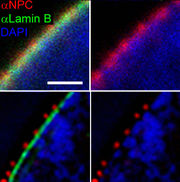
There is also the wide-field structured-illumination (SI) approach to breaking the diffraction limit of light [20][21]. SI—or patterned illumination—relies on both specific microscopy protocols and extensive software analysis post-exposure. But, because SI is a wide-field technique, it is usually able to capture images at a higher rate than confocal-based schemes like STED (This is only a generalization, because SI is not actually superfast). The main concept of SI is to illuminate a sample with patterned light and increase the resolution by measuring the fringes in the Moiré pattern (from the interference of the illumination pattern and the sample). "Otherwise-unobservable sample information can be deduced from the fringes and computationally restored."[22]
SI enhances spatial resolution by collecting information from frequency space outside the observable region. This process is done in reciprocal space: The Fourier transform (FT) of an SI image contains superimposed additional information from different areas of reciprocal space; with several frames with the illumination shifted by some phase, it is possible to computationally separate and reconstruct the FT image, which has much more resolution information. The reverse FT returns the reconstructed image to a super-resolution image.
But this enhances the resolution only by a factor of 2 (because the SI pattern cannot be focused to anything smaller than half the wavelength of the excitation light). To further increase the resolution, one can introduce nonlinearities, which show up as higher-order harmonics in the FT. In reference [22], Gustafsson uses saturation of the fluorescent sample as the nonlinear effect. A sinusoidal saturating excitation beam produces the distorted fluorescence intensity pattern in the emission. This nonpolynomial nonlinearity yields a series of higher-order harmonics in the FT.
Each higher-order harmonic in the FT allows another set of images that can be used to reconstruct a larger area in reciprocal space, and thus a higher resolution. In this case, Gustafsson achieves less than 50-nm resolving power, more than five times that of the microscope in its normal configuration.
The main problems with SI are that, in this incarnation, saturating excitation powers cause more photodamage and lower fluorophore photostability, and sample drift must be kept to below the resolving distance. The former limitation might be solved by using a different nonlinearity (such as stimulated emission depletion or reversible photoactivation, both of which are used in other sub-diffraction imaging schemes); the latter limits live-cell imaging and may require faster frame rates or the use of some fiduciary markers for drift subtraction. Nevertheless, SI is certainly a strong contender for further application in the field of super-resolution microscopy.
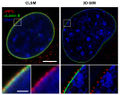 Comparison confocal microscopy - 3D-SIM |
Cell nucleus in prophase from various angles |
 Two mouse cell nuclei in prophase. |
 mouse cell in telophase |
Extensions
Most modern instruments provide simple solutions for micro-photography and image recording electronically. However such capabilities are not always present and the more experienced microscopist will, in many cases, still prefer a hand drawn image rather than a photograph. This is because a microscopist with knowledge of the subject can accurately convert a three dimensional image into a precise two dimensional drawing . In a photograph or other image capture system however, only one thin plane is ever in good focus.
The creation of careful and accurate micrographs requires a microscopical technique using a monocular eyepiece. It is essential that both eyes are open and that the eye that is not observing down the microscope is instead concentrated on a sheet of paper on the bench besides the microscope. With practice, and without moving the head or eyes, it is possible to accurately record the observed details by tracing round the observed shapes by simultaneously "seeing" the pencil point in the microscopical image.
Practicing this technique also establishes good general microscopical technique. It is always less tiring to observe with the microscope focused so that the image is seen at infinity and with both eyes open at all times.
Other enhancements
X-ray
As resolution depends on the wavelength of the light. Electron microscopy has been developed since the 1930s that use electron beams instead of light. Because of the much smaller wavelength of the electron beam, resolution is far higher.
Though less common, X-ray microscopy has also been developed since the late 1940s. The resolution of X-ray microscopy lies between that of light microscopy and the electron microscopy.
Electron microscopy
For light microscopy, the wavelength of the light limits the resolution to around 0.2 micrometers. In order to gain higher resolution, the use of an electron beam with a far smaller wavelength is used in electron microscopes.
- Transmission electron microscopy (TEM) is quite similar to the compound light microscope, by sending an electron beam through a very thin slice of the specimen. The resolution limit in 2005 was around 0.05 nanometer and has not increased appreciably since that time.
- Scanning electron microscopy (SEM) visualizes details on the surfaces of cells and particles and gives a very nice 3D view. It gives results much like those of the stereo light microscope, and, akin to that, its most useful magnification is in the lower range than that of the transmission electron microscope.
Atomic de Broglie
The atomic de Broglie microscope is an imaging system which is expected to provide resolution at the nanometer scale using neutral He atoms as probe particles. [23][24]. Such a device could provide the resolution at nanometer scale and be absolutely non-destructive, but it is not developed as well as optical or electron microscopes.
Scanning probe microscopy
This is a sub-diffraction technique. Examples of scanning probe microscopes are the atomic force microscope (AFM), the Scanning tunneling microscope and the photonic force microscope. All such methods imply a solid probe tip in the vicinity (near field) of an object, which is supposed to be almost flat.
Ultrasonic force
Ultrasonic Force Microscopy (UFM) has been developed in order to improve the details and image contrast on "flat" areas of interest where the AFM images are limited in contrast. The combination of AFM-UFM allows a near field acoustic microscopic image to be generated. The AFM tip is used to detect the ultrasonic waves and overcomes the limitation of wavelength that occurs in acoustic microscopy. By using the elastic changes under the AFM tip, an image of much greater detail than the AFM topography can be generated.
Ultrasonic force microscopy allows the local mapping of elasticity in atomic force microscopy by the application of ultrasonic vibration to the cantilever or sample. In an attempt to analyze the results of ultrasonic force microscopy in a quantitative fashion, a force-distance curve measurement is done with ultrasonic vibration applied to the cantilever base, and the results are compared with a model of the cantilever dynamics and tip-sample interaction based on the finite-difference technique.
Infrared microscopy
The term infrared microscope covers two main types of diffraction-limited microscopy. The first provides optical visualization plus IR spectroscopic data collection. The second (more recent and more advanced) technique employs focal plane array detection for infrared chemical imaging, where the image contrast is determined by the response of individual sample regions to particular IR wavelengths selected by the user.
IR versions of sub-diffraction microscopy (see above) exist also. These include IR NSOM [25] and photothermal microspectroscopy.
Digital holographic microscopy
In digital holographic microscopy (DHM), interfering wave-fronts from a coherent light-source are recorded on a sensor and the image digitally reconstructed by a computer. The image yielded provides a quantitative measurement of the optical thickness of the specimen. DHM can be used with many different optical set-ups. In reflecting DHM, the sensor is positioned on the same side of the specimen as the light source. In transmitting DHM, the sensor and the light source are positioned on opposite sides of the specimen.
One unique feature of DHM is the ability to adjust focus after the image is recorded, since all focus planes are recorded simultaneously by the hologram. The extraction of an individual plane from the three-dimensional hologram data is called sectional image reconstruction.[26]
Holographic snapshots also can be fit to predictions of Lorenz-Mie light scattering theory to track objects' three-dimensional motions with nanometer resolution and simultaneously to measure their size, shape and optical properties. [27]
Digital Pathology (virtual microscopy)
Digital Pathology is an image-based information environment enabled by computer technology that allows for the management of information generated from a digital slide. Digital pathology is enabled in part by virtual microscopy, which is the practice of converting glass slides into digital slides that can be viewed, managed, and analyzed.
Amateur microscopy
Amateur Microscopy is the investigation and observation of biological and non-biological specimens for recreational purposes. Collectors of minerals, insects, seashells, and plants may use microscopes as tools to uncover features that help them classify their collected items. Other amateurs may be interested in observing the life found in pond water and of other samples. Microscopes may also prove useful for the water quality assessment for people that keep a home aquarium. Photographic documentation and drawing of the microscopic images are additional tasks that augment the spectrum of tasks of the amateur. There are even competitions for photomicrograph art. Participants of this pastime either may use commercially prepared microscopic slides or may engage in the task of specimen preparation.
While microscopy is a central tool in the documentation of biological specimens, it is, in general, insufficient to justify the description of a new species based on microscopic investigations alone. Often genetic and biochemical tests are necessary to confirm the discovery of a new species. A laboratory and access to academic literature is a necessity, which is specialized and, in general, not available to amateurs. There is, however, one huge advantage that amateurs have above professionals: time to explore their surroundings. Often, advanced amateurs team up with professionals to validate their findings and (possibly) describe new species.
In the late 1800s, amateur microscopy became a popular hobby in the United States and Europe. Several 'professional amateurs' were being paid for their sampling trips and microscopic explorations by philanthropists, to keep them amused on the Sunday afternoon (e.g., the diatom specialist A. Grunow, being paid by (among others) a Belgian industrialist). Professor John Phin published "Practical Hints on the Selection and Use of the Microscope (Second Edition, 1878)," and was also the editor of the “American Journal of Microscopy.”
In 1995, a loose group of amateur microscopists, drawn from several organizations in the UK and USA, founded a site for microscopy based on the knowledge and input of amateur (perhaps better referred to as 'enthusiast') microscopists. This was the first attempt to establish 'amateur' microscopy as a serious subject in the then-emerging new media of the Internet. Today, it remains as a powerful established international resource for all ages, to input their findings and share information. It is a nonprofit-making web presence dedicated to the pursuit of science and understanding of the small-scale world: [1]
Examples of amateur microscopy images:
 "house bee" Mouth 100X |
 Rice Stem cs 400X |
 Rabbit Testis 100X |
 Fern Porothallium 400X |
See also
- Acronyms in microscopy
- Digital microscope
- Digital Pathology
- Interferometric microscopy
- Köhler illumination
- Timeline of microscope technology
- Two-photon excitation microscopy
References
- ↑ Abramowitz M, Davidson MW (2007). "Introduction to Microscopy". Molecular Expressions. http://micro.magnet.fsu.edu/primer/anatomy/introduction.html. Retrieved 2007-08-22.
- ↑ Abramowitz M, Davidson MW (2003-08-01). "Darkfield Illumination". http://micro.magnet.fsu.edu/primer/techniques/darkfield.html. Retrieved 2008-10-21.
- ↑ Abramowitz M, Davidson MW (2003-08-01). "Rheinberg Illumination". http://micro.magnet.fsu.edu/primer/techniques/rheinberg.html. Retrieved 2008-10-21.
- ↑ Nasse M. J., Woehl J. C. (2010). "Realistic modeling of the illumination point spread function in confocal scanning optical microscopy". J. Opt. Soc. Am. A 27 (2): 295–302. doi:10.1364/JOSAA.27.000295.
- ↑ Wallace W, Schaefer LH, Swedlow JR (2001). "A workingperson's guide to deconvolution in light microscopy". BioTechniques 31 (5): 1076–8, 1080, 1082 passim. PMID 11730015.
- ↑ Considerations on a laser-scanning-microscope with high resolution and depth of field: C. Cremer and T. Cremer in M1CROSCOPICA ACTA VOL. 81 NUMBER 1 September,pp. 31—44 (1978)
- ↑ WEM News and Views
- ↑ Nano-structure analysis using Spatially Modulated Illumination microscopy: D. Baddeley, C. Batram, Y. Weiland, C. Cremer, U.J. Birk in NATURE PROTOCOLS, Vol 2, pp. 2640–2646 (2007)
- ↑ High-precision structural analysis of subnuclear complexes in fixed and live cells via Spatially Modulated Illumination (SMI) microscopy: J. Reymann, D. Baddeley, P. Lemmer, W. Stadter, T. Jegou, K. Rippe, C. Cremer, U. Birk in CHROMOSOME RESEARCH, Vol. 16, pp. 367 –382 (2008)
- ↑ SPDM – Light Microscopy with Single Molecule Resolution at the Nanoscale: P. Lemmer, M.Gunkel, D.Baddeley, R. Kaufmann, A. Urich, Y. Weiland, J.Reymann, P. Müller, M. Hausmann, C. Cremer in APPLIED PHYSICS B, Vol 93, pp. 1-12 (2008).
- ↑ Manuel Gunkel, Fabian Erdel, Karsten Rippe, Paul Lemmer, Rainer Kaufmann, Christoph Hörmann, Roman Amberger and Christoph Cremer: Dual color localization microscopy of cellular nanostructures. In: Biotechnology Journal, 2009, 4, 927-938. ISSN 1860-6768
- ↑ "Fresnel Diffraction Applet" (Java applet). http://www.falstad.com/diffraction/. Retrieved 2007-08-22.
- ↑ Cummings JR, Fellers TJ, Davidson MW (2007). "Specialized Microscopy Techniques - Near-Field Scanning Optical Microscopy". Olympus Microscopy Resource Center. http://www.olympusmicro.com/primer/techniques/nearfield/nearfieldintro.html. Retrieved 2007-08-22.
- ↑ Sánchez EJ, Novotny L, Xie XS (1999). "Near-Field Fluorescence Microscopy Based on Two-Photon Excitation with Metal Tips". Phys Rev Lett 82: 4014–7. doi:10.1103/PhysRevLett.82.4014. http://link.aps.org/abstract/PRL/v82/p4014.
- ↑ Schuck PJ, Fromm DP, Sundaramurthy A, Kino GS, Moerner WE (2005). "Improving the Mismatch between Light and Nanoscale Objects with Gold Bowtie Nanoantennas". Phys Rev Lett 94 (1): 017402. doi:10.1103/PhysRevLett.94.017402. PMID 15698131. http://link.aps.org/abstract/PRL/v94/e017402.
- ↑ STED
- ↑ Webb paper
- ↑ PALM
- ↑ STORM
- ↑ Bailey, B.; Farkas, D. L.; Taylor, D. L.; Lanni, F. Enhancement of axial resolution in fluorescence microscopy by standing-wave excitation. Nature 1993, 366, 44–48.
- ↑ Gustafsson, M. G. L. Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. of Microsc. 2000, 198(2), 82–87.
- ↑ 22.0 22.1 Gustafsson, M. G. L. http://dx.doi.org/10.1073/pnas.0406877102 Nonlinear structured-illumination microscopy: Wide-field fluorescence imaging with theoretically unlimited resolution. PNAS 2005, 102(37), 13081–13086.
- ↑ D.Kouznetsov; H. Oberst, K. Shimizu, A. Neumann, Y. Kuznetsova, J.-F. Bisson, K. Ueda, S. R. J. Brueck (2006). "Ridged atomic mirrors and atomic nanoscope". JOPB 39: 1605–1623. doi:10.1088/0953-4075/39/7/005. http://stacks.iop.org/0953-4075/39/1605.
- ↑ Atom Optics and Helium Atom Microscopy. Cambridge University, http://www-sp.phy.cam.ac.uk/research/mirror.php3
- ↑ H M Pollock and D A Smith, The use of near-field probes for vibrational spectroscopy and photothermal imaging, in Handbook of vibrational spectroscopy, J.M. Chalmers and P.R. Griffiths (eds), John Wiley & Sons Ltd, Vol. 2, pp. 1472 - 1492 (2002)
- ↑ E. Lam; X. Zhang, H. Vo, T.-C. Poon, G. Indebetouw (2009). "Three-dimensional microscopy and sectional image reconstruction using optical scanning holography". Applied Optics 48: H113–H119. doi:10.1364/AO.48.00H113. http://www.opticsinfobase.org/abstract.cfm?URI=ao-48-34-H113.
- ↑ F. C. Cheong; B. Sun, R. Dreyfus, J. Amato-Grill, K. Xiao, L. Dixon, D. G. Grier (2009). "Flow visualization and flow cytometry with holographic video microscopy". Optics Express 17: 13071–13079. doi:10.1364/OE.17.013071. http://www.opticsinfobase.org/oe/abstract.cfm?URI=oe-17-15-13071.
Further reading
- Advanced Light Microscopy vol. 1 Principles and Basic Properties by Maksymilian Pluta, Elsevier (1988)
- Advanced Light Microscopy vol. 2 Specialised Methods by Maksymilian Pluta, Elsevier (1989)
- Introduction to Light Microscopy by S. Bradbury, B. Bracegirdle, BIOS Scientific Publishers (1998)
- Video Microscopy by Shinya Inoue, Plenum Press (1986)
- [2] 1978: Theoretical basis of super resolution 4Pi microscopy & design of a confocal laser scanning fluorescence microscope
- Portraits of life, one molecule at a time, a feature article on sub-diffraction microscopy from the March 1, 2007 issue of Analytical Chemistry
External links
General
- nOOpia Microscopy & photomicrography - A often-updated microscopy and photomicrography blog
- [3] Bwcon award: world´s fastest superresolution microscope as best business idea
- GFP Superresolution (PDF file; 330 kB)
- Olympus Microscopy Resource Center (website critique)
- Nikon MicroscopyU
- Andor Microscopy Techniques - Various techniques used in microscopy.
- Carl Zeiss "Microscopy from the very beginning", a step by step tutorial into the basics of microscopy.
- Microscopy in Detail - A resource with many illustrations elaborating the most common microscopy techniques
- Microscopy Information - Microscopy information and techniques for teachers, educators and enthusiasts.
- WITec SNOM System - NSOM/SNOM and Hybrid Microscopy techniques in combination with AFM, RAMAN, Confocal, Dark-field, DIC & Fluorescence Microscopy techniques.
- Manawatu Microscopy - first known collaboration environment for Microscopy and Image Analysis.
- Audio microscope glossary
- PSF Lab, freeware (for academic use) permitting the calculation of the Point Spread Function in stratified media including polarization effects based on a rigorous vector-based model.
Techniques
- Ratio-metric Imaging Applications For Microscopes Examples of Ratiometric Imaging Work on a Microscope
- Interactive Fluorescence Dye and Filter Database Carl Zeiss Interactive Fluorescence Dye and Filter Database.
- Images formed by simple microscopes - examples of observations with single-lens microscopes.
Organizations
- Royal Microscopical Society (RMS)
- Microscopy Society of America (MSA)
- European Microscopy Society (EMS)
- Non-membership International online organisation (Mic-UK)
|
|||||||||||||||||
|
|||||||||||||||||
[Special:Contributions,is by Jamie Lean]
