Histidine
| L-Histidine | |
|---|---|
 |
 |
|
Histidine
|
|
|
Other names
2-Amino-3-(1H-imidazol-4-yl)propanoic acid
|
|
| Identifiers | |
| CAS number | 71-00-1 |
| PubChem | 773 |
|
SMILES
N[C@@H](Cc1[nH]cnc1)C(O)=O
|
|
| Properties | |
| Molecular formula | C6H9N3O2 |
| Molar mass | 155.15 g mol−1 |
| Except where noted otherwise, data are given for materials in their standard state (at 25 °C, 100 kPa) |
|
| Infobox references | |
Histidine (abbreviated as His or H)[1] is one of the 22 proteinogenic amino acids. In terms of nutrition, histidine is considered an essential amino acid in human infants. After reaching several years of age, humans begin to synthesize it and it thus becomes a non-essential amino acid. Its codons are CAU and CAC.
Histidine was first isolated by German physician Albrecht Kossel in 1896.
Contents |
Chemical properties
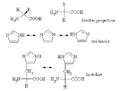
The imidazole sidechain of histidine has a pKa of approximately 6, and, overall, the amino acid has a pKa of 7.6. This means that, at physiologically relevant pH values, relatively small shifts in pH will change its average charge. Below a pH of 6, the imidazole ring is mostly protonated as described by the Henderson–Hasselbalch equation. When protonated, the imidazole ring bears two NH bonds and has a positive charge. The positive charge is equally distributed between both nitrogens and can be represented with two equally important resonance structures.
Aromaticity
The imidazole ring of histidine is aromatic at all pH values. It contains six pi electrons: four from two double bonds and two from a nitrogen lone pair. It can form pi-stacking interactions [2], but is complicated by the positive charge [3]. It doesn't absorb at 280nm in either state, but does in the lower UV range more than some amino acids [4] [5].
Biochemistry
The imidazole sidechain of histidine is a common coordinating ligand in metalloproteins and is a part of catalytic sites in certain enzymes. In catalytic triads, the basic nitrogen of histidine is used to abstract a proton from serine, threonine, or cysteine to activate it as a nucleophile. In a histidine proton shuttle, histidine is used to quickly shuttle protons, it can do this by abstracting a proton with its basic nitrogen to make a positively-charged intermediate and then use another molecule, a buffer, to extract the proton from its acidic nitrogen. In carbonic anhydrases, a histidine proton shuttle is utilized to rapidly shuttle protons away from a zinc-bound water molecule to quickly regenerate the active form of the enzyme.
NMR
As expected, the 15N chemical shifts of these nitrogens are indistinguishable (about 200 ppm, relative to nitric acid on the sigma scale, on which increased shielding corresponds to increased chemical shift). As the pH increases to approximately 8, the protonation of the imidazole ring is lost. The remaining proton of the now neutral imidazole can exist on either nitrogen, giving rise to what are known as the N-1 or N-3 tautomers. NMR shows that the chemical shift of N-1 drops slightly, while the chemical shift of N-3 drops considerably (about 190 vs. 145 ppm). This indicates that the N-1-H tautomer is preferred, presumably due to hydrogen bonding to the neighboring ammonium. The shielding at N-3 is substantially reduced due to the second-order paramagnetic effect, which involves a symmetry-allowed interaction between the nitrogen lone pair and the excited pi* states of the aromatic ring. As the pH rises above 9, the chemical shifts of N-1 and N-3 become approximately 185 and 170 ppm. It is worth noting that the deprotonated form of imidazole, imidazolate ion, would only be formed above a pH of 14, and is therefore not physiologically relevant. This change in chemical shifts can be explained by the presumably decreased hydrogen bonding of an amine over an ammonium ion, and the favorable hydrogen bonding between a carboxylate and an NH. This should act to decrease the N-1-H tautomer preference. [6]
Metabolism
The amino acid is a precursor for histamine and carnosine biosynthesis.

The enzyme histidine ammonia-lyase converts histidine into ammonia and urocanic acid. A deficiency in this enzyme is present in the rare metabolic disorder histidinemia. In Actinobacteria and filamentous fungi, such as Neurospora crassa histidine can be converted into the antioxidant ergothioneine.[7]
Supplementation
Supplementation of Histidine has been shown to cause rapid zinc excretion in rats with an excretion rate 3 to 6 times normal.[8][9]
Additional images
 Histidine |
 The histidine bound heme group of succinate dehydrogenase, an electron carrier in the mitochondrial electron transfer chain. The large semi-transparent sphere indicates the location of the iron ion. From PDB 1YQ3. |
References
- ↑ IUPAC-IUBMB Joint Commission on Biochemical Nomenclature. "Nomenclature and Symbolism for Amino Acids and Peptides". Recommendations on Organic & Biochemical Nomenclature, Symbols & Terminology etc. http://www.chem.qmul.ac.uk/iupac/AminoAcid/. Retrieved 2007-05-17.
- ↑ Lijun Wang,, Na Sun,, Simon Terzyan,, Xuejun Zhang, and, David R. Benson. A Histidine/Tryptophan π-Stacking Interaction Stabilizes the Heme-Independent Folding Core of Microsomal Apocytochrome b5 Relative to that of Mitochondrial Apocytochrome b5. Biochemistry 2006 45 (46), 13750-13759
- ↑ Robert H. Blessing, Edward L. McGandy. Base stacking and hydrogen bonding in crystals of imidazolium dihydrogen orthophosphate. Journal of the American Chemical Society 1972 94 (11), 4034-4035.
- ↑ Katoh R. Absorption Spectra of Imidazolium Ionic Liquids. Chemistry Letters. Vol. 36 (2007) , No. 10 p.1256.
- ↑ AR Goldfarb, LJ Saidel, E Mosovich. THE ULTRAVIOLET ABSORPTION SPECTRA OF PROTEINS. Journal of Biological Chemistry, 1951, p.397-404.
- ↑ Roberts, John D. (2000). ABCs of FT-NMR. Sausalito, CA: University Science Books. pp. 258–259. ISBN 978-1-891389-18-4. http://www.uscibooks.com/.
- ↑ Fahey RC (2001). "Novel thiols of prokaryotes". Annu. Rev. Microbiol. 55: 333–56. doi:10.1146/annurev.micro.55.1.333. PMID 11544359.
- ↑ Freeman, Rm; Taylor, Pr (Apr 1977). "Influence of histidine administration on zinc metabolism in the rat." (Free full text). The American journal of clinical nutrition 30 (4): 523–7. ISSN 0002-9165. PMID 851080. http://www.ajcn.org/cgi/pmidlookup?view=long&pmid=851080.
- ↑ Wensink, J; Van, Den, Hamer, Cj (Jul 1988). "Effect of excess dietary histidine on rate of turnover of 65Zn in brain of rat.". Biological trace element research 16 (2): 137–50. doi:10.1007/BF02797098. PMID 2484542.
See also
- Imidazole
- Aromatic amino acids
- Urocanic aciduria
- Carnosinemia
- Beta-alanine
External links
|
||||||||||||||||||||||||||||||||||
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||