Histone
In biology, histones are strongly alkaline proteins found in eukaryotic cell nuclei, which package and order the DNA into structural units called nucleosomes.[1][2] They are the chief protein components of chromatin, act as spools around which DNA winds, and play a role in gene regulation. Without histones, the unwound DNA in chromosomes would be very long (a length to width ratio of more than 10 million to one in human DNA). For example, each human cell has about 1.8 meters of DNA, but wound on the histones it has about 90 millimeters of chromatin, which, when duplicated and condensed during mitosis, result in about 120 micrometers of chromosomes.[3]
Contents |
Classes
| Core histone H2A/H2B/H3/H4 | |||||
|---|---|---|---|---|---|
| PDB rendering of H2AFJ based on 1aoi. | |||||
| Identifiers | |||||
| Symbol | Histone | ||||
| Pfam | PF00125 | ||||
| Pfam clan | CL0012 | ||||
| InterPro | IPR007125 | ||||
| SCOP | 1hio | ||||
|
|||||
| linker histone H1 and H5 family | |||||
|---|---|---|---|---|---|
 |
|||||
| PDB rendering of HIST1H1B based on 1ghc. | |||||
| Identifiers | |||||
| Symbol | Linker_histone | ||||
| Pfam | PF00538 | ||||
| InterPro | IPR005818 | ||||
| SMART | SM00526 | ||||
| SCOP | 1hst | ||||
|
|||||
Histones "are highly conserved and can be grouped into five major classes: H1/H5, H2A, H2B, H3, and H4".[2][4][5] These are organised into two super-classes as follows:
- core histones – H2A, H2B, H3 and H4
- linker histones – H1 and H5
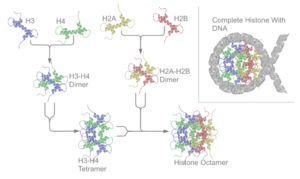
Two of each of the core histones assemble to form one octameric nucleosome core particle by wrapping 147 base pairs of DNA around the protein spool in 1.65 left-handed super-helical turn.[6] The linker histone H1 binds the nucleosome and the entry and exit sites of the DNA, thus locking the DNA into place[7] and allowing the formation of higher order structure. The most basic such formation is the 10 nm fiber or beads on a string conformation. This involves the wrapping of DNA around nucleosomes with approximately 50 base pairs of DNA separating each pair of nucleosomes (also referred to as linker DNA). The assembled histones and DNA is called chromatin. Higher-order structures include the 30 nm fiber (forming an irregular zigzag) and 100 nm fiber, these being the structures found in normal cells. During mitosis and meiosis, the condensed chromosomes are assembled through interactions between nucleosomes and other regulatory proteins.
The following is a list of human histone proteins:
| Super family | Family | Subfamily | Members |
|---|---|---|---|
| Linker | |||
| H1 | |||
| H1F | H1F0, H1FNT, H1FOO, H1FX | ||
| H1H1 | HIST1H1A, HIST1H1B, HIST1H1C, HIST1H1D, HIST1H1E, HIST1H1T | ||
| Core | |||
| H2A | |||
| H2AF | H2AFB1, H2AFB2, H2AFB3, H2AFJ, H2AFV, H2AFX, H2AFY, H2AFY2, H2AFZ | ||
| H2A1 | HIST1H2AA, HIST1H2AB, HIST1H2AC, HIST1H2AD, HIST1H2AE, HIST1H2AG, HIST1H2AI, HIST1H2AJ, HIST1H2AK, HIST1H2AL, HIST1H2AM | ||
| H2A2 | HIST2H2AA3, HIST2H2AC | ||
| H2B | |||
| H2BF | H2BFM, H2BFO, H2BFS, H2BFWT | ||
| H2B1 | HIST1H2BA, HIST1H2BB, HIST1H2BC, HIST1H2BD, HIST1H2BE, HIST1H2BF, HIST1H2BG, HIST1H2BH, HIST1H2BI, HIST1H2BJ, HIST1H2BK, HIST1H2BL, HIST1H2BM, HIST1H2BN, HIST1H2BO | ||
| H2B2 | HIST2H2BE | ||
| H3 | |||
| H3A1 | HIST1H3A, HIST1H3B, HIST1H3C, HIST1H3D, HIST1H3E, HIST1H3F, HIST1H3G, HIST1H3H, HIST1H3I, HIST1H3J | ||
| H3A2 | HIST2H3C | ||
| H3A3 | HIST3H3 | ||
| H4 | |||
| H41 | HIST1H4A, HIST1H4B, HIST1H4C, HIST1H4D, HIST1H4E, HIST1H4F, HIST1H4G, HIST1H4H, HIST1H4I, HIST1H4J, HIST1H4K, HIST1H4L | ||
| H44 | HIST4H4 |
Structure
The nucleosome core is formed of two H2A-H2B dimers and a H3-H4 tetramer, forming two nearly symmetrical halves by tertiary structure (C2 symmetry; one macromolecule is the mirror image of the other).[6] The H2A-H2B dimers and H3-H4 tetramer also show pseudodyad symmetry. The 4 'core' histones (H2A, H2B, H3 and H4) are relatively similar in structure and are highly conserved through evolution, all featuring a 'helix turn helix turn helix' motif (which allows the easy dimerisation). They also share the feature of long 'tails' on one end of the amino acid structure - this being the location of post-translational modification (see below).
Using an electron paramagnetic resonance spin-labeling technique, British researchers measured the distances between the spools around which eukaryotic cells wind their DNA. They determined the spacings range from 59 to 70 Å.[8]
In all, histones make five types of interactions with DNA:
- Helix-dipoles from alpha-helices in H2B, H3, and H4 cause a net positive charge to accumulate at the point of interaction with negatively charged phosphate groups on DNA
- Hydrogen bonds between the DNA backbone and the amide group on the main chain of histone proteins
- Nonpolar interactions between the histone and deoxyribose sugars on DNA
- Salt bridges and hydrogen bonds between side chains of basic amino acids (especially lysine and arginine) and phosphate oxygens on DNA
- Non-specific minor groove insertions of the H3 and H2B N-terminal tails into two minor grooves each on the DNA molecule
The highly basic nature of histones, aside from facilitating DNA-histone interactions, contributes to the water solubility of histones.
Histones are subject to post translational modification by enzymes primarily on their N-terminal tails, but also in their globular domains. Such modifications include methylation, citrullination, acetylation, phosphorylation, SUMOylation, ubiquitination, and ADP-ribosylation. This affects their function of gene regulation (see functions).
In general, genes that are active have less bound histone, while inactive genes are highly associated with histones during interphase. It also appears that the structure of histones has been evolutionarily conserved, as any deleterious mutations would be severely maladaptive.
Function
Compacting DNA strands
Histones act as spools around which DNA winds. This enables the compaction necessary to fit the large genomes of eukaryotes inside cell nuclei: the compacted molecule is 40,000 times shorter than an unpacked molecule.
Chromatin regulation
Histones undergo posttranslational modifications which alter their interaction with DNA and nuclear proteins. The H3 and H4 histones have long tails protruding from the nucleosome which can be covalently modified at several places. Modifications of the tail include methylation, acetylation, phosphorylation, ubiquitination, SUMOylation, citrullination and ADP-ribosylation. The core of the histones H2A, H2B and H3 can also be modified. Combinations of modifications are thought to constitute a code, the so-called "histone code."[9][10] Histone modifications act in diverse biological processes such as gene regulation, DNA repair and chromosome condensation (mitosis).
The common nomenclature of histone modifications is:
- The name of the histone (e.g. H3)
- The single letter amino acid abbreviation (e.g. K for Lysine) and the amino acid position in the protein
- The type of modification (Me: methyl, P: phosphate, Ac: acetyl, Ub: ubiquitin)
So H3K4me1 denotes the monomethylation of the 4th residue (a lysine) from the start (i.e., the N-terminal) of the H3 protein.
Examples of histone modifications in transcription regulation include:
| Type of modification |
Histone | ||||||
|---|---|---|---|---|---|---|---|
| H3K4 | H3K9 | H3K14 | H3K27 | H3K79 | H4K20 | H2BK5 | |
| mono-methylation | activation[11] | activation[12] | activation[12] | activation[12][13] | activation[12] | activation[12] | |
| di-methylation | repression[14] | repression[14] | activation[13] | ||||
| tri-methylation | activation[15] | repression[12] | repression[12] | activation,[13] repression[12] |
repression[14] | ||
| acetylation | activation[15] | activation[15] | |||||
History
Histones were discovered in 1884 by Albrecht Kossel. The word "histone" dates from the late 19th century and is from the German "Histon", of uncertain origin: perhaps from Greek histanai or from histos. Until the early 1990s, histones were dismissed as merely packing material for nuclear DNA. During the early 1990s, the regulatory functions of histones were discovered.[16]
The discovery of the H5 histone appears to date back to 1970's,[17][18] and in classification it has been grouped with H1.[2][4][5]
Conservation across species
Histones are found in the nuclei of eukaryotic cells, and in certain Archaea, namely Euryarchaea, but not in bacteria. Archaeal histones may well resemble the evolutionary precursors to eukaryotic histones. Histone proteins are among the most highly conserved proteins in eukaryotes, emphasizing their important role in the biology of the nucleus.[2]:939 In contrast mature sperm cells largely use protamines to package their genomic DNA, most likely to achieve an even higher packaging ratio.[19]
Core histones are highly conserved proteins, that is, there are very few differences among the amino acid sequences of the histone proteins of different species. Linker histone usually has more than one form within a species and is also less conserved than the core histones.
There are some variant forms in some of the major classes. They share amino acid sequence homology and core structural similarity to a specific class of major histones but also have their own feature that is distinct from the major histones. These minor histones usually carry out specific functions of the chromatin metabolism. For example, histone H3-like CenpA is a histone only associated with the centromere region of the chromosome. Histone H2A variant H2A.Z is associated with the promoters of actively transcribed genes and also involved in the prevention of the spread of silent heterochromatin. Another H2A variant H2A.X binds to the DNA with double strand breaks and marks the region undergoing DNA repair. Histone H3.3 is associated with the body of actively transcribed genes.[20]
See also
- Nucleosome
- Chromatin
- Histone-Modifying Enzymes
- Histone deacetylase
- PRMT4 pathway
- Gene silencing
- Genetics
- Histone methyltransferase
- Histone acetyltransferase
References
- ↑ Youngson, Robert M. (2006). Collins Dictionary of Human Biology. Glasgow: HarperCollins. ISBN 0-00-722134-7.
- ↑ 2.0 2.1 2.2 2.3 Cox, Michael; Nelson, David R.; Lehninger, Albert L (2005). Lehninger Principles of Biochemistry. San Francisco: W.H. Freeman. ISBN 0-7167-4339-6.
- ↑ Redon C, Pilch D, Rogakou E, Sedelnikova O, Newrock K, Bonner W (April 2002). "Histone H2A variants H2AX and H2AZ". Curr. Opin. Genet. Dev. 12 (2): 162–9. doi:10.1016/S0959-437X(02)00282-4. PMID 11893489.
- ↑ 4.0 4.1 Bhasin M, Reinherz EL, Reche PA (2006). "Recognition and classification of histones using support vector machine". J. Comput. Biol. 13 (1): 102–12. doi:10.1089/cmb.2006.13.102. PMID 16472024.
- ↑ 5.0 5.1 Hartl, Daniel L.; Freifelder, David; Snyder, Leon A. (1988). Basic Genetics. Boston: Jones and Bartlett Publishers. ISBN 0-86720-090-1.
- ↑ 6.0 6.1 Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ (September 1997). "Crystal structure of the nucleosome core particle at 2.8 A resolution". Nature 389 (6648): 251–60. doi:10.1038/38444. PMID 9305837. PDB 1AOI
- ↑ Farkas, Daniel (1996). DNA simplified: the hitchhiker's guide to DNA. Washington, D.C: AACC Press. ISBN 0-915274-84-1.
- ↑ Ward R, Bowman A, El-Mkami H, Owen-Hughes T, Norman DG (February 2009). "Long distance PELDOR measurements on the histone core particle". J. Am. Chem. Soc. 131 (4): 1348–9. doi:10.1021/ja807918f. PMID 19138067.
- ↑ Strahl BD, Allis CD (Jan 2000). "The language of covalent histone modifications". Nature 403 (6765): 41–5. doi:10.1038/47412. PMID 10638745.
- ↑ Jenuwein T, Allis CD (Aug 2001). "Translating the histone code". Science 293 (5532): 1074–80. doi:10.1126/science.1063127. PMID 11498575.
- ↑ Benevolenskaya EV (August 2007). "Histone H3K4 demethylases are essential in development and differentiation". Biochem. Cell Biol. 85 (4): 435–43. doi:10.1139/o07-057. PMID 17713579.
- ↑ 12.0 12.1 12.2 12.3 12.4 12.5 12.6 12.7 Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K (May 2007). "High-resolution profiling of histone methylations in the human genome". Cell 129 (4): 823–37. doi:10.1016/j.cell.2007.05.009. PMID 17512414.
- ↑ 13.0 13.1 13.2 Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, Blobel GA, Vakoc CR (April 2008). "DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells". Mol. Cell. Biol. 28 (8): 2825–39. doi:10.1128/MCB.02076-07. PMID 18285465.
- ↑ 14.0 14.1 14.2 Rosenfeld JA, Wang Z, Schones DE, Zhao K, DeSalle R, Zhang MQ (2009). "Determination of enriched histone modifications in non-genic portions of the human genome". BMC Genomics 10: 143. doi:10.1186/1471-2164-10-143. PMID 19335899.
- ↑ 15.0 15.1 15.2 Koch CM, Andrews RM, Flicek P, Dillon SC, Karaöz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, James KD, Lefebvre GC, Bruce AW, Dovey OM, Ellis PD, Dhami P, Langford CF, Weng Z, Birney E, Carter NP, Vetrie D, Dunham I (June 2007). "The landscape of histone modifications across 1% of the human genome in five human cell lines". Genome Res. 17 (6): 691–707. doi:10.1101/gr.5704207. PMID 17567990.
- ↑ Hulton CS, Seirafi A, Hinton JC, Sidebotham JM, Waddell L, Pavitt GD, Owen-Hughes T, Spassky A, Buc H, Higgins CF (Nov 1990). "Histone-like protein H1 (H-NS), DNA supercoiling, and gene expression in bacteria". Cell 63 (3): 631–42. doi:10.1016/0092-8674(90)90458-Q. PMID 2171779.
- ↑ Crane-Robinson C, Dancy SE, Bradbury EM, Garel A, Kovacs AM, Champagne M, Daune M (August 1976). "Structural studies of chicken erythrocyte histone H5". Eur. J. Biochem. 67 (2): 379–88. doi:10.1111/j.1432-1033.1976.tb10702.x. PMID 964248.
- ↑ Aviles FJ, Chapman GE, Kneale GG, Crane-Robinson C, Bradbury EM (August 1978). "The conformation of histone H5. Isolation and characterisation of the globular segment". Eur. J. Biochem. 88 (2): 363–71. doi:10.1111/j.1432-1033.1978.tb12457.x. PMID 689022.
- ↑ Clarke HJ (1992). "Nuclear and chromatin composition of mammalian gametes and early embryos". Biochem. Cell Biol. 70 (10-11): 856–66. doi:10.1139/o92-134. PMID 1297351.
- ↑ Ahmad K, Henikoff S (June 2002). "The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly". Mol. Cell 9 (6): 1191–200. doi:10.1016/S1097-2765(02)00542-7. PMID 12086617.
|
|||||||||||||||||||||||||
External links
- Chromatin, Histones & Cathepsin; PMAP The Proteolysis Map-animation
- Nextbio
- Chromatin Regulation Signaling Pathways