Flagellum
A flagellum (pronounced /fləˈdʒɛləm/, or in plural form: flagella) is a tail-like projection that protrudes from the cell body of certain prokaryotic and eukaryotic cells, and functions in locomotion.[1][2][3] There are some notable differences between prokaryotic and eukaryotic flagella, such as protein composition, structure, and mechanism of propulsion. An example of a flagellated bacterium is the ulcer-causing Helicobacter pylori, which uses multiple flagella to propel itself through the mucus lining to reach the stomach epithelium.[4] An example of a eukaryotic flagellated cell is the sperm cell, which uses its flagellum to propel itself through the female reproductive tract.[5] Eukaryotic flagella are structurally identical to eukaryotic cilia, although distinctions are sometimes made according to function and/or length.[6]
The word flagellum is the Latin word for whip.
Contents |
Types
Three types of flagella have so far been distinguished; bacterial, archaeal and eukaryotic.
The main differences among these three types are summarized below:
- Bacterial flagella are helical filaments that rotate like screws.[7][8][9] They provide two of several kinds of bacterial motility.[10][11]
- Archaeal flagella are superficially similar to bacterial flagella, but are different in many details and considered non-homologous.[12][13][14]
- Eukaryotic flagella - those of animal, plant, and protist cells - are complex cellular projections that lash back and forth.
Sometimes eukaryotic flagella are called cilia or undulipodia to emphasize their distinctiveness.
Bacterial
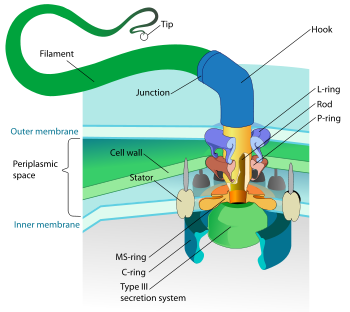


The bacterial flagellum is made up of the protein flagellin. Its shape is a 20 nanometer-thick hollow tube. It is helical and has a sharp bend just outside the outer membrane; this "hook" allows the helix to point directly away from the cell. A shaft runs between the hook and the basal body, passing through protein rings in the cell's membrane that act as bearings. Gram-positive organisms have 2 of these basal body rings, one in the peptidoglycan layer and one in the plasma membrane. Gram-negative organisms have 4 such rings: the L ring associates with the lipopolysaccharides, the P ring associates with peptidoglycan layer, the M ring is embedded in the plasma membrane, and the S ring is directly attached to the plasma membrane. The filament ends with a capping protein.[15][16]
The bacterial flagellum is driven by a rotary engine (the Mot complex) made up of protein, located at the flagellum's anchor point on the inner cell membrane. The engine is powered by proton motive force, i.e., by the flow of protons (hydrogen ions) across the bacterial cell membrane due to a concentration gradient set up by the cell's metabolism (in Vibrio species there are two kinds of flagella, lateral and polar, and some are driven by a sodium ion pump rather than a proton pump[17]). The rotor transports protons across the membrane, and is turned in the process. The rotor alone can operate at 6,000 to 17,000 rpm, but with the flagellar filament attached usually only reaches 200 to 1000 rpm. The direction of rotation can be switched almost instantaneously, caused by a slight change in the position of a protein, FliG, in the rotor[18].
The cylindrical shape of flagella is suited to locomotion of microscopic organisms; these organisms operate at a low Reynolds number, where the viscosity of the surrounding water is much more important than its mass or inertia. [19]
Flagella do not rotate at a constant speed but instead can increase or decrease their rotational speed in relation to the strength of the proton motive force. Flagellar rotation can move bacteria through liquid media at speeds of up to 60 cell lengths/second (sec). Although this is only about 0.00017 km/h (0.00011 mph), when comparing this speed with that of higher organisms in terms of number of lengths moved per second, it is extremely fast. By comparison, the cheetah, the fastest land animal, can sprint at 110 km/h (68 mph), which is approximately 25 body lengths/sec.[20]
During flagellar assembly, components of the flagellum pass through the hollow cores of the basal body and the nascent filament. During assembly, protein components are added at the flagellar tip rather than at the base.[21] In vitro, flagellar filaments assemble spontaneously in a solution containing purified flagellin as the sole protein.
The flagellar filament is the long helical screw that propels the bacterium when rotated by the motor, through the hook. In most bacteria that have been studied, including the Gram negative Escherichia coli, Salmonella typhimurium, Caulobacter crescentus, and Vibrio alginolyticus, the filament is made up of eleven protofilaments approximately parallel to the filament axis. Each protofilament is a series of tandem protein chains. However in Campylobacter jejuni, there are seven protofilaments.[22]
The basal body has several traits in common with some types of secretory pores, such as the hollow rod-like "plug" in their centers extending out through the plasma membrane. Given the structural similarities between bacterial flagella and bacterial secretory systems, it is thought that bacterial flagella may have evolved from the type three secretion system; however, it is not known for certain whether these pores are derived from the bacterial flagella or the bacterial secretory system.
Through use of their flagella, E. coli are able to move rapidly towards attractants and away from repellents. They do this by means of a biased random walk, with 'runs' and 'tumbles' brought about by rotating the flagellum counter-clockwise and clockwise respectively.
Flagella arrangement schemes
Different species of bacteria have different numbers and arrangements of flagella. Monotrichous bacteria have a single flagellum (e.g., Vibrio cholerae). Lophotrichous bacteria have multiple flagella located at the same spot on the bacteria's surfaces which act in concert to drive the bacteria in a single direction. In many cases, the bases of multiple flagella are surrounded by a specialized region of the cell membrane, the so-called polar membrane. Amphitrichous bacteria have a single flagellum on each of two opposite ends (only one flagellum operates at a time, allowing the bacteria to reverse course rapidly by switching which flagellum is active). Peritrichous bacteria have flagella projecting in all directions (e.g., E. coli).
In some bacteria, such as the larger forms of Selenomonas, the individual flagella are organized outside the cell body, helically twining about each other to form a thick structure called a "fascicle". Other bacteria, such as Spirochetes, have a specialized type of flagellum called an "axial filament" that is located in the periplasmic space, the rotation of which causes the entire bacterium to move forward in a corkscrew-like motion.
Counterclockwise rotation of monotrichous polar flagella pushes the cell forward with the flagella trailing behind, much like a corkscrew moving inside cork. Indeed water in the microscopic scale is highly viscous, very different from our daily experience of water. The flagella are left-handed helices, and bundle and rotate together only when rotating counterclockwise. When some of the rotors reverse direction, the flagella unwind and the cell starts "tumbling". It has also been suggested that even if all flagella would rotate clockwise, they will not form a bundle, due to geometrical as well as hydrodynamical reasons.[23][24] Such "tumbling" may happen occasionally, leading to the cell seemingly thrashing about in place, resulting in the reorientation of the cell. The clockwise rotation of a flagellum is suppressed by chemical compounds favorable to the cell (e.g. food), but the motor is highly adaptive to this. Therefore, when moving in a favorable direction, the concentration of the chemical attractant increases and "tumbles" are continually suppressed; however, when the cell's direction of motion is unfavorable (e.g., away from a chemical attractant), tumbles are no longer suppressed and occur much more often, with the chance that the cell will be thus reoriented in the correct direction.
In some Vibrio spp. (particularly Vibrio parahemolyticus[25]) and related proteobacteria such as Aeromonas, two flagellar systems co-exist, using different sets of genes and different ion gradients for energy. The polar flagella are constitutively expressed and provide motility in bulk fluid, while the lateral flagella are expressed when the polar flagella meet too much resistance to turn.[26][27][28][29][30][31] These provide swarming motility on surfaces or in viscous fluids.
Archaeal
The archaeal flagellum is superficially similar to the bacterial (or eubacterial) flagellum; in the 1980s they were thought to be homologous on the basis of gross morphology and behavior.[32] Both flagella consist of filaments extending outside of the cell, and rotate to propel the cell. Archaeal flagella have a unique structure which lacks a central channel. Similar to bacterial type IV pilins, the component flagellins are made with class 3 signal peptides and they are processed by a type IV prepilin peptidase-like enzyme. The archaeal flagellins are typically modified by the addition of N-linked glycans which are necessary for proper assembly and/or function.[3]
Discoveries in the 1990s revealed numerous detailed differences between the archaeal and bacterial flagella; these include:
- Bacterial flagella are motorized by a flow of H+ ions (or occasionally Na+ ions); archaeal flagella are almost certainly powered by ATP. The torque-generating motor that powers rotation of the archaeal flagellum has not been identified.
- While bacterial cells often have many flagellar filaments, each of which rotates independently, the archaeal flagellum is composed of a bundle of many filaments that rotate as a single assembly.
- Bacterial flagella grow by the addition of flagellin subunits at the tip; archaeal flagella grow by the addition of subunits to the base.
- Bacterial flagella are thicker than archaeal flagella, and the bacterial filament has a large enough hollow "tube" inside that the flagellin subunits can flow up the inside of the filament and get added at the tip; the archaeal flagellum is too thin to allow this.
- Many components of bacterial flagella share sequence similarity to components of the type III secretion systems, but the components of bacterial and archaeal flagella share no sequence similarity. Instead, some components of archaeal flagella share sequence and morphological similarity with components of type IV pili, which are assembled through the action of type II secretion systems (the nomenclature of pili and protein secretion systems is not consistent).
These differences could mean that the bacterial and archaeal flagella could be a classic case of biological analogy, or convergent evolution, rather than homology. However, in comparison to the decades of well-publicized study of bacterial flagella (e.g. by Berg), archaeal flagella have only recently begun to get serious scientific attention. Therefore, many assume erroneously that there is only one basic kind of prokaryotic flagellum, and that archaeal flagella are homologous to it. For example, Cavalier-Smith (2002)[32] is aware of the differences between archaeal and bacterial flagellins, but retains the misconception that the basal bodies are homologous.
Eukaryotic



Along with cilia, flagella make up a group of organelles known as undulipodia.
Structure
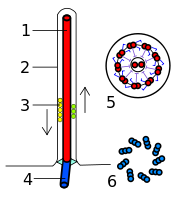
A eukaryotic flagellum is a bundle of nine fused pairs of microtubule doublets surrounding two central single microtubules. The so-called "9+2" structure is characteristic of the core of the eukaryotic flagellum called an axoneme. At the base of a eukaryotic flagellum is a basal body, "blepharoplast" or kinetosome, which is the microtubule organizing center (MTOC) for flagellar microtubules and is about 500 nanometers long. Basal bodies are structurally identical to centrioles. The flagellum is encased within the cell's plasma membrane, so that the interior of the flagellum is accessible to the cell's cytoplasm.
Mechanism
Each of the outer 9 doublet microtubules extends a pair of dynein arms (an "inner" and an "outer" arm) to the adjacent microtubule; these dynein arms are responsible for flagellar beating, as the force produced by the arms causes the microtubule doublets to slide against each other and the flagellum as a whole to bend. These dynein arms produce force through ATP hydrolysis. The flagellar axoneme also contains radial spokes, polypeptide complexes extending from each of the outer 9 microtubule doublets towards the central pair, with the "head" of the spoke facing inwards. The radial spoke is thought to be involved in the regulation of flagellar motion, although its exact function and method of action are not yet understood.
Flagella vs Cilia

Though eukaryotic flagella and motile cilia are ultrastructurally identical, the beating pattern of the two organelles can be different. In the case of flagella (e.g. the tail of a sperm) the motion is propeller-like. In contrast, beating of motile cilia consists of coordinated back-and-forth cycling of many cilia on the cell surface. Thus, flagella serve for the propulsion of single cells (e.g. swimming of protozoa and spermatozoa), and motile cilia for the transport of fluids (e.g. transport of mucus by stationary ciliated cells in the trachea). However, cilia are also used for locomotion (through liquids) in organisms such as Paramecium.
Intraflagellar Transport
Intraflagellar transport (IFT), the process by which axonemal subunits, transmembrane receptors, and other proteins are moved up and down the length of the flagellum, is essential for proper functioning of the flagellum, in both motility and signal transduction.[33]
For information on biologists' ideas about how the various flagella may have evolved, see evolution of flagella.
Evolution of flagella and debate
Based on similarity in structure and partial similarity in amino acid sequence, it is generally accepted among scientists that the eukaryotic flagellum and cilium have evolved from the cytoskeleton, while the eubacterial flagellum has evolved either from the type III secretion system or from a more ancient secretion system from which the type III secretion system has evolved as well. The archaeal flagellum has probably evolved from the type IV pili. More details appear under Evolution of flagella.
In his 1996 book Darwin's Black Box, intelligent design proponent Michael Behe, under funding from the Templeton Foundation, cited the bacterial flagellum as an example of an irreducibly complex structure that could not have evolved through naturalistic means. Behe argued that the flagellum becomes useless if any one of its constituent parts is removed, and thus could not have arisen through numerous, successive, slight modifications; therefore, it is hopelessly improbable that the proteins making up the flagellar motor could have come together all at once, by chance.[34] Mark Perakh explained that while Behe popularized the idea, biologist Hermann J. Müller had already explored it (under the slightly different name of “interlocking complexity”) and more than a decade before Behe’s book the same idea was explored by A. Graham Cairns-Smith, but neither claimed that “irreducible complexity” was a “marker” of a supernatural design.[35]
While Behe discussed the immune system and the blood clotting cascade in greater detail, the bacterial flagellum has become a "poster child" for intelligent design proponents and other creationists, as during the Kitzmiller v. Dover Area School District trial of 2005.[36] It is one of two identified rotary structures found in nature (the other being ATP synthase)[37] and it is billions of years older than Behe's other two examples, which exist in many homologous forms, simplifying the explanation of their origin.[38]
Evolutionary pathways supported by the Theory of Natural Selection and Evolution (see: "The Flagellum Unspun and PBS/Nova Science's television production of Intelligent Design on Trial) have since been identified for the bacterial flagellum; thus, undermining Behe's argument.[39] In addition, the Type three secretion system, a molecular syringe which bacteria use to inject toxins into other cells, appears to be a simplified sub-set of the bacterial flagellum's components, meaning that it is much less likely to be irreducibly complex in the way that the bacterial flagellum could have in fact evolved from the type three secretion system.[40][41]
Exaptation explains how systems with multiple parts can evolve through natural means.[42]
See also
- Cilia
- Evolution of flagella
- Genetic ciliopathy
- Rotation in living systems
- Undulipodia
References
- ↑ Bardy SL, Ng SY, Jarrell KF (February 2003). "Prokaryotic motility structures". Microbiology (Reading, Engl.) 149 (Pt 2): 295–304. doi:10.1099/mic.0.25948-0. PMID 12624192.
- ↑ Lefebvre PA; Lefebvre, PA (2001). "Assembly and Motility of Eukaryotic Cilia and Flagella. Lessons from Chlamydomonas reinhardtii". Plant Physiol. 127 (4): 1500–1507. doi:10.1104/pp.010807. PMID 11743094.
- ↑ 3.0 3.1 Jarrell, K (editor) (2009). Pili and Flagella: Current Research and Future Trends. Caister Academic Press. ISBN 978-1-904455-48-6.
- ↑ Lacy BE, Rosemore J (October 2001). "Helicobacter pylori: ulcers and more: the beginning of an era" (abstract page). J. Nutr. 131 (10): 2789S–2793S. PMID 11584108. http://jn.nutrition.org/cgi/content/abstract/131/10/2789S.
- ↑ Malo AF, Gomendio M, Garde J, Lang-Lenton B, Soler AJ, Roldan ER (June 2006). "Sperm design and sperm function". Biol. Lett. 2 (2): 246–9. doi:10.1098/rsbl.2006.0449. PMID 17148374.
- ↑ Haimo LT, Rosenbaum JL (December 1981). "Cilia, flagella, and microtubules". J. Cell Biol. 91 (3 Pt 2): 125s–130s. doi:10.1083/jcb.91.3.125s. PMID 6459327.
- ↑ Silverman M, Simon M (1974). "Flagellar rotation and the mechanism of bacterial motility". Nature 249 (452): 73–74. doi:10.1038/249073a0. PMID 4598030.
- ↑ Meister GLM, Berg HC (1987). "Rapid rotation of flagellar bundles in swimming bacteria". Nature 325: 637–640. doi:10.1038/325637a0.
- ↑ Berg HC, Anderson RA (1973). "Bacteria Swim by Rotating their Flagellar Filaments". Nature 245 (5425): 380–382. doi:10.1038/245380a0. PMID 4593496.
- ↑ Jahn TL, Bovee EC (1965). "Movement and Locomotion of Microorganisms". Annual Review of Microbiology 19: 21–58. doi:10.1146/annurev.mi.19.100165.000321. PMID 5318439.
- ↑ Harshey RM (2003). "Bacterial Motility on a Surface: Many Ways to a Common Goal". Annual Review of Microbiology 57: 249–273. doi:10.1146/annurev.micro.57.030502.091014. PMID 14527279.
- ↑ Ng SY, Chaban B, Jarrell KF (2006). "Archaeal flagella, bacterial flagella and type IV pili: a comparison of genes and posttranslational modifications". J. Mol. Microbiol. Biotechnol. 11 (3–5): 167–91. doi:10.1159/000094053. PMID 16983194.
- ↑ Metlina AL (2004). "Bacterial and archaeal flagella as prokaryotic motility organelles". Biochemistry Mosc. 69 (11): 1203–12. doi:10.1007/s10541-005-0065-8. PMID 15627373.
- ↑ Jarrell et al (2009). "Archaeal Flagella and Pili". Pili and Flagella: Current Research and Future Trends. Caister Academic Press. ISBN 978-1-904455-48-6.
- ↑ Macnab RM (2003). "How bacteria assemble flagella". Annu. Rev. Microbiol. 57: 77–100. doi:10.1146/annurev.micro.57.030502.090832. PMID 12730325.
- ↑ Diószeghy Z, Závodszky P, Namba K, Vonderviszt F (2004). "Stabilization of flagellar filaments by HAP2 capping". FEBS Lett. 568 (1–3): 105–9. doi:10.1016/j.febslet.2004.05.029. PMID 15196929.
- ↑ Atsumi T, McCarter L, Imae Y. (1992). "Polar and lateral flagellar motors of marine Vibrio are driven by different ion-motive forces". Nature 355 (6356): 182–4. doi:10.1038/355182a0. PMID 1309599.
- ↑ Dean, Tim. "Inside nature’s most efficient motor: the flagellar", Australian Life Scientist, 02 August 2010. Retrieved on 2010-08-04.
- ↑ Dusenbery DB (2009). "Chapter 13". Living at Micro Scale: The Unexpected Physics of Being Small. Cambridge: Harvard University Press. ISBN 0-674-03116-4.
- ↑ Milton Hildebrand (1959). "Motions of Cheetah and Horse". Journal of Mammalogy. http://www.jstor.org/view/00222372/ap050163/05a00030/12?frame=noframe&userID=83e69956@siu.edu/01cce4405a00501cdc4b9&dpi=3&config=jstor. Retrieved 2007-10-30. Although according to Cheetah, Luke Hunter and Dave Hamman, (Struik Publishers, 2003), pp. 37–38, the cheetah's fastest recorded speed was 110 km/h (68 mph).
- ↑ Minamino T, Imada K, Namba K. (2008). "Mechanisms of type III protein export for bacterial flagellar assembly". Mol. Biosyst. 4 (11): 1105–15. doi:10.1039/b808065h. PMID 18931786.
- ↑ Galkin VE, Yu X, Bielnicki J, Heuser J, Ewing CP, Guerry P, Egelman EH. (2008). "Divergence of quaternary structures among bacterial flagellar filaments.". Science 320 (5874): 382–5. doi:10.1126/science.1155307. PMID 18420936.
- ↑ Kim M, Bird JC, Van Parys AJ, Breuer KS, Powers TR (December 2003). "A macroscopic scale model of bacterial flagellar bundling". Proc. Natl. Acad. Sci. U.S.A. 100 (26): 15481–5. doi:10.1073/pnas.2633596100. PMID 14671319.
- ↑ Macnab RM (January 1977). "Bacterial flagella rotating in bundles: a study in helical geometry". Proc. Natl. Acad. Sci. U.S.A. 74 (1): 221–5. doi:10.1073/pnas.74.1.221. PMID 264676.
- ↑ Kim YK, McCarter LL (2000). "Analysis of the Polar Flagellar Gene System of Vibrio parahaemolyticus". Journal of Bacteriology 182 (13): 3693–3704. doi:10.1128/JB.182.13.3693-3704.2000. PMID 10850984. PMC 94540. http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=10850984.
- ↑ Atsumi T, Maekawa Y, Yamada T, Kawagishi I, Imae Y, Homma M (1 August 1996). "Effect of viscosity on swimming by the lateral and polar flagella of Vibrio alginolyticus". Journal of Bacteriology 178 (16): 5024–5026. PMID 8759871. PMC 178290. http://jb.asm.org/cgi/pmidlookup?view=long&pmid=8759871.
- ↑ McCarter LL (2004). "Dual Flagellar Systems Enable Motility under Different Circumstances". Journal of Molecular Microbiology and Biotechnology 7 (1-2): 18–29. doi:10.1159/000077866. PMID 15170400.
- ↑ Merino S, Shaw JG, Tomás JM. (2006). "Bacterial lateral flagella: an inducible flagella system". FEMS Microbiol Lett 263 (2): 127–35. doi:10.1111/j.1574-6968.2006.00403.x. PMID 16978346.
- ↑ Belas R, Simon M, Silverman M. (1986). "Regulation of lateral flagella gene transcription in Vibrio parahaemolyticus". J Bacteriol 167 (1): 210–8. PMID 3013835. PMC 212863. http://jb.asm.org/cgi/content/abstract/167/1/210.
- ↑ Canals R, Altarriba M, Vilches S, Horsburgh G, Shaw JG, Tomás JM, Merino S (2006). "Analysis of the Lateral Flagellar Gene System of Aeromonas hydrophila AH-3". Journal of Bacteriology 188 (3): 852–862. doi:10.1128/JB.188.3.852-862.2006. PMID 16428388.
- ↑ Canals R, Ramirez S, Vilches S, Horsburgh G, Shaw JG, Tomás JM, Merino S (January 2006). "Polar flagellum biogenesis in Aeromonas hydrophila". J. Bacteriol. 188 (2): 542–55. doi:10.1128/JB.188.2.542-555.2006. PMID 16385045.
- ↑ 32.0 32.1 Cavalier-Smith T (1987). "The origin of eukaryotic and archaebacterial cells". Ann. N. Y. Acad. Sci. 503: 17–54. doi:10.1111/j.1749-6632.1987.tb40596.x. PMID 3113314. http://www.annalsnyas.org/cgi/content/citation/503/1/17.
- ↑ Pazour GJ (October 2004). "Intraflagellar transport and cilia-dependent renal disease: the ciliary hypothesis of polycystic kidney disease". J. Am. Soc. Nephrol. 15 (10): 2528–36. doi:10.1097/01.ASN.0000141055.57643.E0. PMID 15466257.
- ↑ Behe, Michael (1996.) Darwin's Black Box: The Biochemical Challenge to Evolution. New York: Free Press. (p. 70–73) as quoted in The Bacterial Flagellum as an example of irreducible complexity. ARN Molecular Museum (retrieved 4 January 2008.)
- ↑ Mark Perakh (2006-02-25). "Bacteria Flagella Look Like Man-made Machines". Skeptic (U.S. magazine). http://www.skeptic.com/the_magazine/. Retrieved 2006-11-06.
- ↑ 40 Days and 40 Nights: Darwin, Intelligent Design, God, OxyContin, and Other Oddities on Trial in Pennsylvania. (Harper Collins, April 10, 2007) ISBN 0061179450
- ↑ Yoshida M, Muneyuki E, Hisabori T (September 2001). "ATP synthase--a marvellous rotary engine of the cell". Nat. Rev. Mol. Cell Biol. 2 (9): 669–77. doi:10.1038/35089509. PMID 11533724. http://stahlberglab.ucdavis.edu/teaching/mcb-221/pdf/yoshida-natrevm-2001-atpase.pdf. Retrieved 2008-06-02.
- ↑ Matzke NJ (2003). "Background to "Evolution in (Brownian) space". TalkOrigins.org. http://www.talkdesign.org/faqs/flagellum_background.html. Retrieved 2008-06-02.
- ↑ Pallen MJ, Matzke NJ (October 2006). "From The Origin of Species to the origin of bacterial flagella". Nat. Rev. Microbiol. 4 (10): 784–90. doi:10.1038/nrmicro1493. PMID 16953248.
- ↑ Miller KR. "The Flagellum Unspun: The Collapse of "Irreducible Complexity"". www.millerandlevine.com. http://www.millerandlevine.com/km/evol/design2/article.html. Retrieved 2008-06-02.
- ↑ Dembski WA (2003-02-17). "The bacterial flagellum: still spinning just fine". Design Inference Website. http://www.designinference.com/documents/2003.02.Miller_Response.htm. Retrieved 2008-06-02.
- ↑ "We therefore find that Professor Behe’s claim for irreducible complexity has been refuted in peer-reviewed research papers and has been rejected by the scientific community at large." Ruling, Judge John E. Jones III, Kitzmiller v. Dover Area School District
External links
- "Molecular Machines Museum Index". Access Research Network. 2001. http://www.arn.org/mm/mm_index.htm. Retrieved 2008-05-18.
- Howard Berg (1999). "Motile Behavior of Bacteria". Physics Today on the Web. American Institute of Physics. http://www.aip.org/pt/jan00/berg.htm. Retrieved 2008-05-18.
- Charles Lindemann (2008-04-04). "Mechanisms of sperm motility". Oakland University. http://www2.oakland.edu/biology/lindemann/. Retrieved 2008-05-18.
E.M. Purcell (1977). "Life at Low Reynolds Number". American Journal of Physics vol 45. pp. 3–11. http://www.nada.kth.se/~annak/Low_RE.pdf.
|
|||||||||||||||||||||||||||||||||||
|
||||||||||||||||||||||||||
|
|||||||||||||||||||||||||
![]() This article incorporates content from the 1728 Cyclopaedia, a publication in the public domain.
This article incorporates content from the 1728 Cyclopaedia, a publication in the public domain.

