Ubiquitin
Ubiquitin is a highly-conserved regulatory protein that is ubiquitously expressed in eukaryotes. Ubiquitination (or ubiquitylation) refers to the post-translational modification of a protein by the covalent attachment (via an isopeptide bond) of one or more ubiquitin monomers. The most prominent function of ubiquitin is labeling proteins for proteasomal degradation. Besides this function, ubiquitination also controls the stability, function, and intracellular localization of a wide variety of proteins.
Contents |
Identification
Ubiquitin (originally, Ubiquitous Immunopoietic Polypeptide) was first identified in 1975 as an 8.5-kDa protein of unknown function expressed universally in living cells. The basic functions of ubiquitin and the components of the ubiquitination pathway were elucidated in the early 1980s in groundbreaking work performed by Aaron Ciechanover, Avram Hershko, and Irwin Rose for which the Nobel Prize in Chemistry was awarded in 2004.[1]
The ubiquitylation system was initially characterised as an ATP-dependent proteolytic system present in cellular extracts. A heat-stable polypeptide present in these extracts, ATP-dependent proteolysis factor 1 (APF-1), was found to become covalently attached to the model protein substrate lysozyme in an ATP- and Mg2+-dependent process. Multiple APF-1 molecules were linked to a single substrate molecule by an isopeptide linkage, and conjugates were found to be rapidly degraded with the release of free APF-1. Soon after APF-1-protein conjugation was characterised, APF-1 was identified as ubiquitin. The carboxyl group of the C-terminal glycine residue of ubiquitin (Gly76) was identified as the moiety conjugated to substrate lysine residues.
The protein
| Number of residues | 76 |
| Molecular mass | 8564.47 Da |
| Isoelectric point (pI) | 6.79 |
| Gene names | RPS27A (UBA80, UBCEP1), UBA52 (UBCEP2), UBB, UBC |
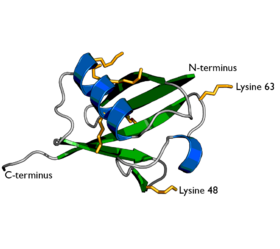
Ubiquitin is a small protein that occurs in all eukaryotic cells. It performs its myriad functions through conjugation to a large range of target proteins. A variety of different modifications can occur. The ubiquitin protein itself consists of 76 amino acids and has a molecular mass of about 8.5 kDa. Key features include its C-terminal tail and the 7 Lys residues. It is highly conserved among eukaryotic species: Human and yeast ubiquitin share 96% sequence identity. The human ubiquitin sequence in one-letter code (lysine residues in bold):
| MQIFVKTLTGKTITLEVEPSDTIENVKAKIQDKEGIPPDQQRLIFAGKQLEDGRTLSDYNIQKESTLHLVLRLRGG |
Ubiquitination (Ubiquitylation)

The process of marking a protein with ubiquitin (ubiquitylation or ubiquitination) consists of a series of steps:
- Activation of ubiquitin: Ubiquitin is activated in a two-step reaction by an E1 ubiquitin-activating enzyme in a process requiring ATP as an energy source. The initial step involves production of a ubiquitin-adenylate intermediate. The second step transfers ubiquitin to the E1 active site cysteine residue, with release of AMP. This step results in a thioester linkage between the C-terminal carboxyl group of ubiquitin and the E1 cysteine sulfhydryl group.
- Transfer of ubiquitin from E1 to the active site cysteine of a ubiquitin-conjugating enzyme E2 via a trans(thio)esterification reaction. Mammalian genomes contain 30-40 UBCs.
- The final step of the ubiquitylation cascade creates an Isopeptide bond between a lysine of the target protein and the C-terminal glycine of ubiquitin. In general, this step requires the activity of one of the hundreds of E3 ubiquitin-protein ligases (often termed simply ubiquitin ligase). E3 enzymes function as the substrate recognition modules of the system and are capable of interaction with both E2 and substrate. E3 enzymes possess one of two domains:
- The HECT (Homologous to the E6-AP Carboxyl Terminus) domain
- The RING (Really Interesting New Gene) domain (or the closely-related U-box domain)
- Transfer can occur in two ways:
-
- Directly from E2, catalysed by RING domain E3s.
- Via an E3 enzyme, catalysed by HECT domain E3s. In this case, a covalent E3-ubiquitin intermediate is formed before transfer of ubiquitin to the substrate protein.
-
Multisubunit E3 ubiquitin ligases
The Anaphase-promoting complex (APC) and the SCF complex (for Skp1-Cullin-F-box protein complex) are two examples of multi-subunit E3s involved in recognition and ubiquitination of specific target proteins for degradation by the proteasome.
Function and variety of ubiquitin modifications
Following addition of a single ubiquitin moiety to a protein substrate (monoubiquitination), further ubiquitin molecules can be added to the first, yielding a polyubiquitin chain. In addition, some substrates are modified by addition of ubiquitin molecules to multiple lysine residues in a process termed multiubiquitination. As discussed, ubiquitin possesses a total of 7 lysine residues. Historically the original type of ubiquitin chains identified were those linked via lysine 48. However, more recent work has uncovered a wide variety of linkages involving all possible lysine residues[2] and in addition chains assembled on the N-terminus of a ubiquitin molecule ("linear chains")[3]. Work published in 2007 has demonstrated the formation of branched ubiquitin chains containing multiple linkage types[4]. "Atypical" (non-lysine 48-linked) ubiquitin chains have been discussed in a review by Ikeda & Dikic[5]
The ubiquitination system functions in a wide variety of cellular processes, including[6]:
- Antigen processing
- Apoptosis
- Biogenesis of organelles
- Cell cycle and division
- DNA transcription and repair
- Differentiation and development
- Immune response and inflammation
- Neural and muscular degeneration
- Morphogenesis of neural networks
- Modulation of cell surface receptors, ion channels and the secretory pathway
- Response to stress and extracellular modulators
- Ribosome biogenesis
- Viral infection
Lysine 48-linked chains

The most studied polyubiquitin chains - lysine48-linked - target proteins for destruction, a process known as proteolysis. At least four ubiquitin molecules must be attached to a lysine residue on the condemned protein in order for it to be recognised by the 26S-proteasome[7]. The proteasome is a complex, barrel-shaped structure with two chambers, within which proteolysis occurs. Proteins are rapidly degraded into small peptides (usually 3-24 amino acid residues in length). Ubiquitin molecules are cleaved off the protein immediately prior to destruction and are recycled for further use. Although the majority of proteasomal substrates are ubiquitinated, there are examples of non-ubiquitinated proteins being targeted to the proteasome.
Lysine 63-linked chains

Monoubiquitination
Ubiquitin can also mark transmembrane proteins (for example, receptors) for removal from membranes and fulfill several signaling roles within the cell. Cell-surface transmembrane molecules that are tagged with ubiquitin are often monoubiquitinated, and this modification alters the subcellular localization of the protein, often targeting the protein for destruction in lysosomes.
Other chain types
Disease association
Genetic disorders
Some genetic disorders associated with ubiquitin are:
- The gene whose disruption causes Angelman syndrome, UBE3A, encodes a ubiquitin ligase (E3) enzyme termed E6-AP.
- The gene disrupted in Von Hippel-Lindau syndrome encodes a ubiquitin E3 ligase termed the VHL tumor suppressor or VHL gene.
- The gene disrupted in Liddle's Syndrome results in disregulation of an epithelial Na+ channel (ENaC) and causes hypertension.
- Eight of the thirteen identified genes whose disruption causes Fanconi anemia encode proteins that form a large ubiquitin ligase (E3) complex.
- mutations of the Cullin7 E3 ubiquitin ligase are linked to 3-M syndrome, an autosomal-recessive growth retardation disorder[8]
Immunohistochemistry
Antibodies to ubiquitin are used in histology to identify abnormal accumulations of protein inside cells that are markers of disease. These accumulations are called inclusion bodies. Examples of such abnormal inclusions in cells are
- Neurofibrillary tangles in Alzheimer's disease
- Lewy body in Parkinson's disease
- Pick bodies in Pick's disease
- Inclusions in motor neuron disease and Huntington's Disease
- Mallory bodies in alcoholic liver disease
- Rosenthal fibers in astrocytes
Ubiquitin-like modifiers
Although ubiquitin is the most well understood post-translation modifier, there is a growing family of ubiquitin-like proteins (UBLs) that modify cellular targets in a pathway that is parallel to but distinct from that of ubiquitin. These alternative modifiers include: SUMO (Sentrin, Smt3 in yeast), NEDD8 (Rub1 in yeast), ISG15 (UCRP), APG8, APG12, FAT10, Ufm1 URM1 & Hub1.
These related molecules have novel functions and influence diverse biological processes. There is also cross-regulation between the various conjugation pathways since some proteins can become modified by more than one UBL, and sometimes even at the same lysine residue. For instance, SUMO modification often acts antagonistically to that of ubiquitination and serves to stabilize protein substrates. Proteins conjugated to UBLs are typically not targeted for degradation by the proteasome, but rather function in diverse regulatory activities. Attachment of UBLs might alter substrate conformation, affect the affinity for ligands or other interacting molecules, alter substrate localization and influence protein stability.
UBLs are structurally similar to ubiquitin and are processed, activated, conjugated and released from conjugates by enzymatic steps that are similar to the corresponding mechanisms for ubiquitin. UBLs are also translated with C-terminal extensions that are processed to expose the invariant C-terminal LRGG. These modifiers have their own specific E1 (activating), E2 (conjugating) and E3 (ligating) enzymes that conjugate the UBLs to intracellular targets. These conjugates can be reversed by UBL-specific isopeptidases that have similar mechanisms to that of the deubiquitinating enzymes.[9]
References
- ↑ "Official website of Nobel Prize Commitee, list of 2004 winners". Retrieved on 04/30/2008.
- ↑ Xu, Ping; Junmin Peng (2008-05-01). "Characterization of polyubiquitin chain structure by middle-down mass spectrometry". Analytical Chemistry 80 (9): 3438-44. doi:. http://www.ncbi.nlm.nih.gov/pubmed/18351785. Peng, Junmin; Daniel Schwartz, Joshua E Elias, Carson C Thoreen, Dongmei Cheng, Gerald Marsischky, Jeroen Roelofs, Daniel Finley, Steven P Gygi (2003-08). "A proteomics approach to understanding protein ubiquitination". Nature Biotechnology 21 (8): 921-6. doi:. http://www.ncbi.nlm.nih.gov/pubmed/12872131.
- ↑ Kirisako, Takayoshi; Kiyoko Kamei, Shigeo Murata, Michiko Kato, Hiromi Fukumoto, Masato Kanie, Soichi Sano, Fuminori Tokunaga, Keiji Tanaka, Kazuhiro Iwai (2006-10-18). "A ubiquitin ligase complex assembles linear polyubiquitin chains". The EMBO Journal 25 (20): 4877-87. doi:. http://www.ncbi.nlm.nih.gov/pubmed/17006537.
- ↑ Kim, Hyoung Tae; Kwang Pyo Kim, Fernando Lledias, Alexei F Kisselev, K Matthew Scaglione, Dorota Skowyra, Steven P Gygi, Alfred L Goldberg (2007-06-15). "Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages". The Journal of Biological Chemistry 282 (24): 17375-86. doi:. http://www.ncbi.nlm.nih.gov/pubmed/17426036.
- ↑ Ikeda, Fumiyo; Ivan Dikic (2008-06). "Atypical ubiquitin chains: new molecular signals. 'Protein Modifications: Beyond the Usual Suspects' review series". EMBO Reports 9 (6): 536-42. doi:. http://www.ncbi.nlm.nih.gov/pubmed/18516089.
- ↑ "Ubiquitin Proteasome Pathway Overview". Retrieved on 04/30/2008.
- ↑ Thrower, J S; L Hoffman, M Rechsteiner, C M Pickart (2000-01-04). "Recognition of the polyubiquitin proteolytic signal". The EMBO Journal 19 (1): 94-102. doi:. http://www.ncbi.nlm.nih.gov/pubmed/10619848.
- ↑ Huber, Céline; Dora Dias-Santagata, Anna Glaser, James O'Sullivan, Raja Brauner, Kenneth Wu, Xinsong Xu, Kerra Pearce, Rong Wang, Maria Luisa Giovannucci Uzielli, Nathalie Dagoneau, Wassim Chemaitilly, Andrea Superti-Furga, Heloisa Dos Santos, André Mégarbané, Gilles Morin, Gabriele Gillessen-Kaesbach, Raoul Hennekam, Ineke Van der Burgt, Graeme C M Black, Peter E Clayton, Andrew Read, Martine Le Merrer, Peter J Scambler, Arnold Munnich, Zhen-Qiang Pan, Robin Winter, Valérie Cormier-Daire (2005-10). "Identification of mutations in CUL7 in 3-M syndrome". Nature Genetics 37 (10): 1119-24. doi:. http://www.ncbi.nlm.nih.gov/pubmed/16142236.
- ↑ "Ubiquitin Proteasome Pathway Overview". Retrieved on 04/30/2008.
See also
- Autophagy
- Autophagin
- ERAD
- SUMO protein
- SUMO enzymes
- SUMO network
- Autophagy network
- Ubiquitin ligase
Note: Ubiquitin is also used to mark paternal mitochondria for destruction during human fertilization.
External links
Academic
- UniProt entry for ubiquitin
- UbiPred — server for the prediction of ubiquitylation sites
- Ubiquitin Web-page
Commercial
- Boston Biochem : Ubiquitin Proteasome System Research and Reagents
- UPP Reference Lit : Ubiquitin Proteasome Pathway research paper reference literature list
- Invitrogen : Drug discovery assays
- Progenra Inc. : Ubiquitin-based Biotechnology Corporation
Further reading
- Essays in Biochemistry, Volume 41 (2005): The Ubiquitin-Proteasome System (Portland Press)
|
||||||||||||||||||||||||||||||||||||||||||||