Cladistics
| Part of the Biology series on |
| Evolution |
 |
| Introduction |
| Mechanisms and processes |
|
Adaptation |
| Research and history |
|
Evidence |
| Evolutionary biology fields |
|
Cladistics |
| Biology Portal · |
Cladistics is the hierarchical classification of species based on evolutionary ancestry. Cladistics is distinguished from other taxonomic systems because it focuses on evolution rather than similarities between species, and because it places heavy emphasis on objective, quantitative analysis. Cladistics generates diagrams called cladograms that represent the evolutionary tree of life. DNA and RNA sequencing data are used in many important cladistic efforts. Computer programs are widely used in cladistics, due to the highly complex nature of cladogram generation procedures. Cladistics originated in the work of the German entomologist, Willi Hennig, who himself referred to it as phylogenetic systematics; the use of the terms "cladistics" and "clade" was popularized by other researchers.[1] The term phylogenetics is often used synonymously with cladistics. Cladistics originated in the field of biology but in recent years has found application in other disciplines. The word cladistics is derived from the ancient Greek κλάδος, klados, "branch."
Contents |
Terminology


- A clade is an ancestor species and all of its descendents
- A monophyletic group is a clade
- A paraphyletic group is a monophyletic group that excludes some of the descendants (e.g. reptiles are sauropsids excluding birds). Most cladists discourage the use of paraphyletic groups.
- A polyphyletic group is a group consisting of members from two non-overlapping monophyletic groups (e.g. flying animals). Most cladists discourage the use of polyphyletic groups.
- An outgroup is an organism that is considered not to be part of the group in question, but is closely related to the group.
- A characteristic that is present in both the outgroups and in the ancestors is called a plesiomorphy (meaning "close form", also called an ancestral state).
- A characteristic that occurs only in later descendants is called an apomorphy (meaning "separate form", also called a "derived" state) for that group. Note: The adjectives plesiomorphic and apomorphic are used instead of "primitive" and "advanced" to avoid placing value-judgments on the evolution of the character states, since both may be advantageous in different circumstances. It is not uncommon to refer informally to a collective set of plesiomorphies as a ground plan for the clade or clades they refer to.
- A species or clade is basal to another clade if it holds more plesiomorphic characters than that other clade. Usually a basal group is very species-poor as compared to a more derived group. It is not a requirement that a basal group be extant. For example, palaeodicots are basal to flowering plants.
- A clade or species located within another clade is said to be nested within that clade.
Three definitions of clade
There are three ways to define a clade for use in a cladistic taxonomy.[2]
- Node-based: the most recent common ancestor of A and B, and all its descendants. See crown group.
- Stem-based: all descendants of the oldest common ancestor of A and B that is not also an ancestor of Z. See total group.
- Apomorphy-based: the most recent common ancestor of A and B, along with all of its descendants, possessing a certain derived character. This definition is generally discouraged by most cladists.
History of cladistics
Hennig's major book, even the 1979 version, does not contain the term cladistics in the index. He referred to his own approach as phylogenetic systematics, as implied by the book's title. A review paper by Dupuis observes that the term clade was introduced in 1958 by Julian Huxley, cladistic by Cain and Harrison in 1960, and cladist (for an adherent of Hennig's school) by Mayr in 1965.[3]
From the time of Hennig's original formulation until the end of the 1980s cladistics remained a minority approach to classification. However in the 1990s it rapidly became the dominant method of classification in evolutionary biology. Cheap but increasingly powerful personal computers made it possible to process large quantities of data about organisms and their characteristics. At about the same time the development of effective polymerase chain reaction techniques made it possible to apply cladistic methods of analysis to biochemical features of organisms as well as to anatomical ones.[4]
Cladistics as a successor to phenetics
For some decades in the mid to late 20th century, a commonly used methodology was phenetics ("numerical taxonomy"). This can be seen as a predecessor[5] to some methods of today's cladistics (namely distance matrix methods like neighbor-joining), but made no attempt to resolve phylogeny, only similarities.
Cladograms
The starting point of cladistic analysis is a group of species and molecular, morphological, or other data characterizing those species. The end result is a tree-like relationship diagram called a cladogram,[6] or sometimes a dendrogram (Greek for "tree drawing").[7] The cladogram graphically represents a hypothetical evolutionary process. Cladograms are subject to revision as additional data become available.
Synonyms
The terms evolutionary tree, and sometimes phylogenetic tree are often used synonymously with cladogram,[8] but others treat phylogenetic tree as a broader term that includes trees generated with a nonevolutionary emphasis.
Subtrees are clades
In cladograms, all organisms lie at the leaves.[9] The two taxa on either side of a split are called sister taxa or sister groups. Each subtree, whether it contains only two or a hundred thousand items, is called a clade.
2-way versus 3-way forks
Many cladists require that all forks in a cladogram be 2-way forks. Some cladograms include 3-way or 4-way forks when there are insufficient data to resolve the forking to a higher level of detail. See phylogenetic tree for more information about forking choices in trees.
Number of distinct cladograms
For a given set of species, the number of distinct cladograms that can be drawn (ignoring which cladogram best matches the species characteristics) is:[10]
| Number of Species | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | N |
| Number of Cladograms | 1 | 3 | 15 | 105 | 945 | 10,395 | 135,135 | 2,027,025 | 34,459,425 | 1*3*5*7*...*(2N-3) |
This exponential growth of the number of possible cladograms explains why manual creation of cladograms becomes very difficult when the number of species is large.
Depth
If a cladogram represents N species, the number of levels (the "depth") in the cladogram is on the order of log2(N).[11] For example, if there are 32 species of deer, a cladogram representing deer will be around 5 levels deep (because 25 = 32). A cladogram representing the complete tree of life, with about 10 million species, would be about 23 levels deep. This formula gives a lower limit: in most cases the actual depth will be a larger value because the various branches of the cladogram will not be uniformly deep. Conversely, the depth may be shallower if forks larger than 2-way forks are permitted.
Time scale
A cladogram tree has an implicit time axis,[12] with time running forward from the base of the tree to the leaves of the tree. If the approximate date (for example, expressed as millions of years ago) of all the evolutionary forks were known, those dates could be captured in the cladogram. Thus, the time axis of the cladogram could be assigned a time scale (e.g. 1 cm = 1 million years), and the forks of the tree could be graphically located along the time axis. Such cladograms are called scaled cladograms. Many cladograms are not scaled along the time axis, for a variety of reasons:
- Many cladograms are built from species characteristics that cannot be readily dated (e.g. morphological data in the absence of fossils or other dating information)
- When the characteristic data are DNA/RNA sequences, it is feasible to use sequence differences to establish the relative ages of the forks, but converting those ages into actual years requires a significant approximation of the rate of change[13]
- Even when the dating information is available, positioning the cladogram's forks along the time axis in proportion to their dates may cause the cladogram to become difficult to understand or hard to fit within a human-readable format
Extinct species
Cladistics makes no distinction between extinct and nonextinct species,[14] and it is appropriate to include extinct species in the group of organisms being analyzed. Cladograms that are based on DNA/RNA generally do not include extinct species because DNA/RNA samples from extinct species are rare. Cladograms based on morphology, especially morphological characteristics that are preserved in fossils, are more likely to include extinct species.
Cladistics in taxonomy
Cladistics contrasted with traditional taxonomy

Prior to the advent of cladistics, most taxonomists used Linnaean taxonomy and later Evolutionary taxonomy to organize life forms. These traditional approaches, still in use by some researchers (especially in works intended for a more general audience[16]) use several fixed levels of a hierarchy, such as kingdom, phylum, class, order, and family. Cladistics does not use those terms, because one of the fundamental premises of cladistics is that the evolutionary tree is so deep and so complex that it is inadvisable to set a fixed number of levels.
Evolutionary taxonomy insists that groups reflect phylogenies. In contrast, Linnean taxonomy allows both monophyletic and paraphyletic groups as taxa. Since the early 20th century, Linnaean taxonomists have generally attempted to make genus-level and lower-level taxa monophyletic. Ernst Mayr drew a distinction between the terms cladistics and phylogeny, using the term cladistics to refer to classifications which only take into account genealogy, as opposed to phylogeny, which had previously been used in a broader sense to refer to the combination of genealogy and amount of divergence from an ancestor (i.e. Evolutionary taxonomy). Mayr wrote, in 1985:
It would seem to me to be quite evident that the two concepts of phylogeny (and their role in the construction of classifications) are sufficiently different to require terminological distinction. The term phylogeny should be retained for the broad concept of phylogeny, promoted by Darwin and adopted by most students of phylogeny in the ensuing 90 years. The concept of phylogeny as mere genealogy should be terminologically distinguished as cladistics. To lump the two concepts together terminologically could not help but produce harmful equivocation.
—[17]
Willi Hennig's pioneering work provoked a spirited debate[18] about the relative merits of cladistics versus traditional taxonomy which has continued down to the present.[19] Some of the debates that the cladists engaged in had been running since the 19th century, but they entered these debates with a new fervor,[20] as can be seen from the Foreword to Hennig (1979) by Rosen, Nelson, and Patterson:
Encumbered with vague and slippery ideas about adaptation, fitness, biological species and natural selection, neo-Darwinism (summed up in the "evolutionary" systematics of Mayr and Simpson) not only lacked a definable investigatory method, but came to depend, both for evolutionary interpretation and classification, on consensus or authority.
—Foreword, page ix
Cladistics strictly and exclusively follows phylogeny and has arbitrarily deep trees with binary branching: each taxon is a clade. Linnaean taxonomy, while following phylogeny, also subjectively considers morphology and has a fixed hierarchy, whose taxa are not always clades.
Paraphyletic groups discouraged
Many cladists discourage the use of paraphyletic groups because they detract from cladistics' emphasis on clades (monophyletic groups). In contrast, proponents of the use of paraphyletic groups argue that any dividing line in a cladogram creates both a monophyletic section above and a paraphyletic section below. They also contend that paraphyletic taxa are necessary for classifying earlier sections of the tree – for instance, the early vertebrates that would someday evolve into the family Hominidae cannot be placed in any other monophyletic family. They also argue that paraphyletic taxa provide information about significant changes in organisms' morphology, ecology, or life history – in short, that both paraphyletic groups and clades are valuable notions with separate purposes.
Complexity of the Tree of Life
One argument in favor of cladistics is that it supports arbitrarily complex, arbitrarily deep trees. Especially when extinct species are considered (both known and unknown), the complexity and depth of the tree can be very large. Every single speciation event, including all the species that are now extinct, represents an additional fork on the hypothetical, complete cladogram representing the full tree of life. Fractals can be used to represent this notion of increasing detail: as a viewpoint zooms into the tree of life, the complexity remains virtually constant[21]. This great complexity of the tree, and the uncertainty associated with the complexity, are among the reasons that cladists cite for the attractiveness of cladistics over traditional taxonomy.
Proponents of noncladistic approaches to taxonomy point to punctuated equilibrium to bolster the case that the tree of life has a finite depth and finite complexity. If the number of species currently alive is finite, and the number of extinct species that we will ever know about is finite, then the depth and complexity of the tree of life is bounded, and there is no need to handle arbitrarily deep trees.
PhyloCode approach to naming species
A formal code of phylogenetic nomenclature, the PhyloCode[22], is currently under development for cladistic taxonomy. It is intended for use by both those who would like to abandon Linnaean taxonomy and those who would like to use taxa and clades side by side. In several instances (see for example Hesperornithes) it has been employed to clarify uncertainties in Linnaean systematics so that in combination they yield a taxonomy that unambiguously places problematic groups in the evolutionary tree in a way that is consistent with current knowledge.
Example
-
For more details on this topic, see Reptilia#History of classification.
For example, Linnaean taxonomy contains the taxon Tetrapoda, defined morphologically as vertebrates with four limbs (as well as animals with four-limbed ancestors, such as snakes), which is often given the rank of superclass, and divides into the classes Amphibia, Reptilia, Aves, Mammalia, and some extinct families.
Cladistics also contains the taxon Tetrapoda, whose living members can be classified phylogenically as "the clade defined by the common ancestor of amphibians and mammals", or more precisely the clade defined by the common ancestor of a specific amphibian and mammal (or bird or reptile), but whose tree is still being worked out (there are a number of extinct branches). The taxon does not have a rank, and its subtaxa are subclades: these can be contained within one another, but one does not divide the clade into several non-overlapping taxa (as in traditional taxonomy): one can split into two clades at the first branching, but that is all. With regards to the traditional classes, Aves and Mammalia are subclades, contained in the subclade Amniota, but Reptilia* is a paraphyletic taxon, not a clade — "At best, the cladists suggest, we could say that the traditional Reptilia are "non-avian, non-mammalian amniotes"[23] — and instead one divides Amniota into the two clades Sauropsida (which contains birds and all living amniotes other than mammals, including all living traditional reptiles) and Theropsida (mammals and the extinct "mammal-like reptiles"). Similarly, Amphibia* is a paraphyletic taxon.
Summary of advantages of cladistics
Proponents of cladistics enumerate key distinctions between cladistics and Linnaean taxonomy as follows:[24]
| Cladistics | Linnaean Taxonomy |
| Treats all levels of the tree as equivalent. | Treats each tree level uniquely. Uses special names (such as Family, Class, Order) for each level. |
| Handles arbitrarily deep trees. | Often must invent new level names (such as superorder, suborder, infraorder, parvorder, magnorder) to accommodate new discoveries. Biased towards trees about 4 to 12 levels deep. |
| Discourages naming or use of groups that are not monophyletic | Acceptable to name and use paraphyletic groups |
| Primary goal is to reflect actual process of evolution | Primary goal is to group species based on morphological similarities |
| Assumes that the shape of the tree will change frequently, with new discoveries | New discoveries often require renaming or releveling of Classes, Orders, and Kingdoms |
| Definitions of taxa are objective, hence free from personal interpretation | Definitions of taxa require individuals to make subjective decisions. For example, various taxonomists suggest that the number of Kingdoms in Biology is two, three, four, five, or six. |
| Taxa, once defined, are permanent (e.g. "taxon X comprises the most recent common ancestor of species A and B along with its descendants") | Taxa can be renamed and eliminated (e.g. Aschelminthes and Insectivora are some of many taxa in the Linnaean system that have been eliminated). |
Summary of criticisms of cladistics
Critics of cladistics include Ashlock,[25] Mayr,[26] Williams[27] and Envall[28]. Some of their criticisms include:
| Cladistics | Linnaean Taxonomy |
| Limited to entities related by evolution or ancestry | Supports groupings without reference to evolution or ancestry |
| Does not include a process for naming species | Includes a process for giving unique names to species |
| Difficult to understand the essence of a clade, because clade definitions emphasize ancestry at the expense of meaningful characteristics | Taxa definitions based on tangible characteristics |
| Ignores sensible, clearly defined paraphyletic groups such as reptiles | Permits clearly defined groups such as reptiles |
| Difficult to determine if a given species is in a clade or not (e.g. if clade X is defined as "most recent common ancestor of A and B along with its descendants", then the only way to determine if species Y is in the clade is to perform a complex evolutionary analysis) | Straightforward process to determine if a given species is in a taxon or not |
| Limited to organisms that evolved by inherited traits; not applicable to organisms that evolved via complex gene sharing or lateral transfer | Applicable to all organisms, regardless of evolutionary mechanism |
Process to generate a cladogram

A simplified procedure for generating a cladogram is:[30]
- Gather and organize data
- Consider possible cladograms
- Select best cladogram
Step 1
A cladistic analysis begins with the following data:
- a list of species to be organized
- a list of characteristics to be compared
- for each species, the value of each of the listed characteristics or character states
For example, if analyzing 20 species of birds, the data might be:
- the list of 20 species
- characteristics such as genome sequence, skeletal anatomy, biochemical processes, and feather coloration
- for each of the 20 species, its particular genome sequence, skeletal anatomy, biochemical processes, and feather coloration
Molecular versus morphological data
The characteristics used to create a cladogram can be roughly categorized as either morphological (synapsid skull, warm blooded, notochord, unicellular, etc.) or molecular (DNA, RNA, or other genetic information).[30] Prior to the advent of DNA sequencing, all cladistic analysis used morphological data.
As DNA sequencing has become cheaper and easier, molecular systematics has become a more and more popular way to reconstruct phylogenies.[31] Using a parsimony criterion is only one of several methods to infer a phylogeny from molecular data; maximum likelihood and Bayesian inference, which incorporate explicit models of sequence evolution, are non-Hennigian ways to evaluate sequence data. Another powerful method of reconstructing phylogenies is the use of genomic retrotransposon markers, which are thought to be less prone to the problem of reversion that plagues sequence data. They are also generally assumed to have a low incidence of homoplasies because it was once thought that their integration into the genome was entirely random; this seems at least sometimes not to be the case, however.
Ideally, morphological, molecular, and possibly other phylogenies should be combined into an analysis of total evidence: All have different intrinsic sources of error. For example, character convergence (homoplasy) is much more common in morphological data than in molecular sequence data, but character reversions that are unrecognizable as such are more common in the latter (see long branch attraction). Morphological homoplasies can usually be recognized as such if character states are defined with enough attention to detail.
Plesiomorphies and synapomorphies
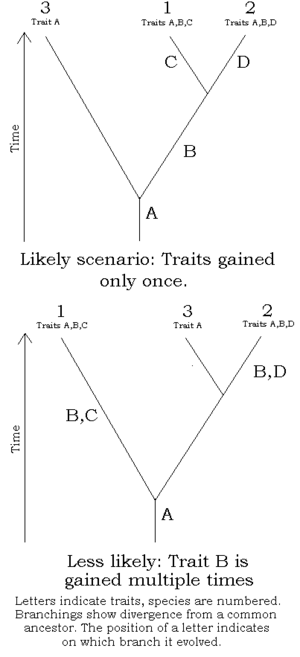
The researcher must decide which character states were present before the last common ancestor of the species group (plesiomorphies) and which were present in the last common ancestor (synapomorphies) and does so by comparison to one or more outgroups. The choice of outgroup is a crucial step in cladistic analysis because different outgroups can produce trees with profoundly different topologies. Note that only synapomorphies are of use in characterizing clades.
Avoid homoplasies
A homoplasy is a character that is shared by multiple species due to some cause other than common ancestry.[32] Typically, homoplasies occur due to convergent evolution. Use of homoplasies when building a cladogram is sometimes unavoidable but is to be avoided when possible.
A well known example of homoplasy due to convergent evolution would be the character, "presence of wings". Though the wings of birds, bats, and insects serve the same function, each evolved independently, as can be seen by their anatomy. If a bird, bat, and a winged insect were scored for the character, "presence of wings", a homoplasy would be introduced into the dataset, and this would confound the analysis, possibly resulting in a false evolutionary scenario.
Homoplasies can often be avoided outright in morphological datasets by defining characters more precisely and increasing their number. When analyzing "supertrees" (datasets incorporating as many taxa of a suspected clade as possible), it may become unavoidable to introduce character definitions that are imprecise, as otherwise the characters might not apply at all to a large number of taxa; to continue with the "wings" example, the presence of wings would hardly be a useful character if attempting a phylogeny of all Metazoa, as most of these don't have wings at all. Cautious choice and definition of characters thus is another important element in cladistic analyses. With a faulty outgroup or character set, no method of evaluation is likely to produce a phylogeny representing the evolutionary reality.
Step 2
When there are just a few species being organized, it is possible to do this step manually, but most cases require a computer program. There are scores of computer programs available to support cladistics.[33] See phylogenetic tree for more information about tree-generating computer programs.
Because the total number of possible cladograms grows exponentially with the number of species, it is impractical for a computer program to evaluate every individual cladogram. A typical cladistic program begins by using heuristic techniques to identify a small number of candidate cladograms. Many cladistic programs then continue the search with the following repetitive steps:
- Evaluate the candidate cladograms by comparing them to the characteristic data
- Identify the best candidates that are most consistent with the characteristic data
- Create additional candidates by creating several variants of each of the best candidates from the prior step
- Use heuristics to create several new candidate cladograms unrelated to the prior candidates
- Repeat these steps until the cladograms stop getting better
Computer programs that generate cladograms use algorithms that are very computationally intensive,[34] because the cladogram algorithm is NP-hard.
Step 3
There are several algorithms available to identify the "best" cladogram.[35] Most algorithms use a metric to measure how consistent a candidate cladogram is with the data. Most cladogram algorithms use the mathematical techniques of optimization and minimization.
In general, cladogram generation algorithms must be implemented as computer programs, although some algorithms can be performed manually when the data sets are trivial (for example, just a few species and a couple of characteristics).
Some algorithms are useful only when the characteristic data are molecular (DNA, RNA); other algorithms are useful only when the characteristic data are morphological. Other algorithms can be used when the characteristic data includes both molecular and morphological data.
Algorithms for cladograms include least squares, neighbor-joining, parsimony, maximum likelihood, and Bayesian inference.
Biologists sometimes use the term parsimony for a specific kind of cladogram generation algorithm and sometimes as an umbrella term for all cladogram algorithms.[36]
Algorithms that perform optimization tasks (such as building cladograms) can be sensitive to the order in which the input data (the list of species and their characteristics) is presented. Inputting the data in various orders can cause the same algorithm to produce different "best" cladograms. In these situations, the user should input the data in various orders and compare the results.
Using different algorithms on a single data set can sometimes yield different "best" cladograms, because each algorithm may have a unique definition of what is "best".
Because of the astronomical number of possible cladograms, algorithms cannot guarantee that the solution is the overall best solution. A nonoptimal cladogram will be selected if the program settles on a local minimum rather than the desired global minimum.[37] To help solve this problem, many cladogram algorithms use a simulated annealing approach to increase the likelihood that the selected cladogram is the optimal one.[38]
Application to other disciplines

The processes used to generate cladograms are not limited to the field of biology[39]. The generic nature of cladistics means that cladistics can be used to organize groups of items in many different academic realms. The only requirement is that the items have characteristics that can be identified and measured.
Recent attempts in the use of cladistic methods outside of biology attack problems in:
- anthropology[40];
- languages;
- the filiation of manuscripts in textual criticism, usually called stemma,[41][42] and
- the lineage of Linux distros, a class of computer operating system.
Footnotes
- ↑ Phylogenetic Systematics is the title of Hennig's 1966 book
- ↑ de Queiroz, K. and J. Gauthier (1994). "Toward a phylogenetic system of biological nomenclature". Trends in Research in Ecology and Evolution 9 (1): 27–31. doi:.
- ↑ Dupuis, Claude (1984). "Willi Hennig's impact on taxonomic thought". Annual Review of Ecology and Systematics 15: 1–24. ISSN 0066-4162.
- ↑ Baron, C., and Høeg, J.T., (2005). ""Gould, Scharm and the paleontologocal perspective in evolutionary biology"". in Koenemann, S., and Jenner, R.A.. Crustacea and Arthropod Relationships. CRC Press. pp. 3–14. ISBN 0849334985. http://books.google.co.uk/books?id=LalmQ4346O0C&dq=Nielsen,+C.+2001+%22Animal+evolution%22+Chelicerata&source=gbs_summary_s&cad=0. Retrieved on 2008-10-15.
- ↑ Mayr, Ernst (1982). The growth of biological thought: diversity, evolution and inheritance. Cambridge, MA: Harvard Univ. Press. p. 221. ISBN 0-674-36446-5.
- ↑ pp. 45, 78 and 555 of Joel Cracraft and Michael J. Donaghue, eds. (2004). Assembling the Tree of Life. Oxford, England: Oxford University Press.
- ↑ Weygoldt, P. (February 1998). "Evolution and systematics of the Chelicerata". Experimental and Applied Acarology 22 (2): 63–79. doi:.
- ↑ Singh, Gurcharan (2004). Plant Systematics: An Integrated Approach. Science. pp. 203–4. ISBN 1578083516.
- ↑ Albert, Victor (2006). Parsimony, Phylogeny, and Genomics. Oxford University Press. p. 3-55. ISBN 0199297304.
- ↑ Lowe, Andrew (2004). Ecological Genetics: Design, Analysis, and Application. Blackwell Publishing. p. 164. ISBN 1405100338.
- ↑ Aldous, David (1996), "Probability Distributions on Cladograms", Random Discrete Structures, Springer, pp. 13
- ↑ Freeman, Scott (1998). Evolutionary Analysis. Prentice Hall. p. 380. ISBN 0135680239.
- ↑ Carroll, Robert Lynn (1997). Patterns and Processes of Vertebrate Evolution. Cambridge University Press. p. 80. ISBN 052147809X.
- ↑ Scott-Ram, N. R. (1990). Transformed Cladistics, Taxonomy and Evolution. Cambridge University Press. p. 83. ISBN 0521340861.
- ↑ Letunic, I (2007). "Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation" (Pubmed). Bioinformatics 23(1): 127–8. doi:. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=pubmed&dopt=Abstract&list_uids=17050570.
- ↑ Unwin, David M. (2006). The Pterosaurs: From Deep Time. New York: Pi Press. pp. 246. ISBN ISBN 0-13-146308-X.
- ↑ Mayr, E. (1985). "Darwin and the Definition of Phylogeny." Systematic Zoology, 34(1): 97-98.
- ↑ Wheeler, Quentin (2000). Species Concepts and Phylogenetic Theory: A Debate. Columbia University Press. ISBN 0231101430.
- ↑ Benton, M. (2000). "Stems, nodes, crown clades, and rank-free lists: is Linnaeus dead?". Biological Reviews 75 (4): 633–648.
- ↑ Hull, David (1988). Science as a Process. University of Chicago Press. p. 232-276. ISBN 0226360512.
- ↑ Gordon, Richard (1999). The Hierarchical Genome and Differentiation Waves. World Scientific. p. 632. ISBN 9810222688.
- ↑ Pennisi, E. (2001). "Evolutionary Biology: Preparing the Ground for a Modern 'Tree of Life'". Science 293: 1979–1980. doi:.
- ↑ Colin Tudge (2000). The Variety of Life. Oxford University Press. ISBN 0198604262.
- ↑ Hennig, Willi (1975). "'Cladistic analysis or cladistic classification': a reply to Ernst Mayr". Systematic Zoology 24: 244–256. doi:.
- ↑ Ashlock PD. 1971. Monophyly and associated terms. Systematic Zoology 20: 63–69.
Ashlock PD. 1972. Monophyly again. Systematic Zoology 21: 430–438.
Ashlock PD. 1974. The uses of cladistics. Annual Review of Ecology and Systematics 5: 81–89.
Ashlock PD. 1979. An evolutionary systematist’s view of classification. Systematic Zoology 28: 441–450. - ↑ Mayr E. 1974. Cladistic analysis or cladistic classification? Zeitschrift fűr Zoologische Systematik und Evolutionforschung 12: 94–128.
Mayr E. 1978. Origin and history of some terms in systematic and evolutionary biology. Systematic Zoology 27: 83–88.
Mayr E, Bock WJ. 2002. Classifications and other ordering systems. Journal of Zoological Systematics and Evolutionary Research 40: 169–194. - ↑ Williams, P.A. 1992. Confusion in cladism. Synthese 01:135-132
- ↑ Envall, M. 2008. On the difference between mono-, holo- and paraphyletic groups - a consistent distinction of process and pattern. Biol. J. Linn. Soc. 94:217-220.
- ↑ Hodge T, Cope M (2000). "A myosin family tree". J Cell Sci 113 Pt 19: 3353–4. PMID 10984423. http://jcs.biologists.org/cgi/content/full/113/19/3353.
- ↑ 30.0 30.1 DeSalle, Rob (2002). Techniques in Molecular Systematics and Evolution. Birkhauser. ISBN 376436257X.
- ↑ Hillis, David (1996). Molecular Systematics. Sinaur. ISBN 0878932828.
- ↑ West-Eberhard, Mary (2003). Developmental Plasticity and Evolution. Oxford Univ. Press. pp. 353–376. ISBN 0195122356.
- ↑ "List of Cladistics Software Programs".
- ↑ Hodkinson, Trevor (2006). Reconstructing the Tree of Life: Taxonomy and Systematics of Species Rich Taxa. CRC Press. p. 61-128. ISBN 0849395798.
- ↑ Kitching, Ian (1998). Cladistics: The Theory and Practice of Parsimony Analysis. Oxford University Press. ISBN 0198501382.
- ↑ Stewart, Caro-Beth (1993). "The Powers and Pitfalls of Parsimony". Nature 361: 603–607. doi:.
- ↑ Foley, Peter (1993). Cladistics: A Practical Course in Systematics. Oxford Univ. Press. p. 66. ISBN 0198577664.
- ↑ Nixon K. C. (1999). "The Parsimony Ratchet: a new method for rapid parsimony analysis". Cladistics 15: 407–414. doi:.
- ↑ Mace, Ruth (2005). The Evolution of Cultural Diversity: A Phylogenetic Approach. Routledge Cavendish. ISBN 1844720993.
- ↑ Lipo, Carl (2005). Mapping Our Ancestors: Phylogenetic Approaches in Anthropology and Prehistory. Aldine Transaction. ISBN 0202307514.
- ↑ Metzger & Ehrman, The text of the New Testament, OUP, 2005, p.207f.
- ↑ Peter M.W. Robinson, & Robert J. O’Hara. 1996. Cladistic analysis of an Old Norse manuscript tradition. Research in Humanities Computing, 4: 115–137. Available at http://rjohara.net/cv/1996-rhc.
See also
|
|
|
References
- Ashlock, Peter D. (1974). "The uses of cladistics". Annual Review of Ecology and Systematics 5: 81–99. doi:. ISSN 0066-4162.
- Cuénot, Lucien (1940). "Remarques sur un essai d'arbre généalogique du règne animal". Comptes Rendus de l'Académie des Sciences de Paris 210: 23–27. Available free online at http://gallica.bnf.fr (No direct URL). This is the paper credited by Hennig (1979) for the first use of the term 'clade'.
- Cavalli-Sforza, L.L. and A.W.F. Edwards (Sep., 1967). "Phylogenetic analysis: Models and estimation procedures". Evol. 21 (3): 550–570. doi:. http://links.jstor.org/sici?sici=0014-3820%28196709%2921%3A3%3C550%3APAMAEP%3E2.0.CO%3B2-I.
- de Queiroz, Kevin (1992). "Phylogenetic taxonomy". Annual Review of Ecology and Systematics 23: 449–480. doi:. ISSN 0066-4162.
- Dupuis, Claude (1984). "Willi Hennig's impact on taxonomic thought". Annual Review of Ecology and Systematics 15: 1–24. ISSN 0066-4162.
- Felsenstein, Joseph (2004). Inferring phylogenies. Sunderland, MA: Sinauer Associates. ISBN 0-87893-177-5.
- Hamdi, Hamdi; Hitomi Nishio, Rita Zielinski and Achilles Dugaiczyk (1999). "Origin and phylogenetic distribution of Alu DNA repeats: irreversible events in the evolution of primates". Journal of Molecular Biology 289: 861–871. doi:. PMID 10369767.
- Hennig, Willi (1950). Grundzüge einer Theorie der Phylogenetischen Systematik. Berlin: Deutscher Zentralverlag..
- Hennig, Willi (1982). Phylogenetische Systematik (ed. Wolfgang Hennig). Berlin: Blackwell Wissenschaft. ISBN 3-8263-2841-8.
- Hennig, Willi (1975). "'Cladistic analysis or cladistic classification': a reply to Ernst Mayr". Systematic Zoology 24: 244–256. doi:. The paper he was responding to is reprinted in Mayr (1976).
- Hennig, Willi (1966). Phylogenetic systematics (tr. D. Dwight Davis and Rainer Zangerl). Urbana, IL: Univ. of Illinois Press (reprinted 1979 and 1999). ISBN 0-252-06814-9.
- Hennig, Willi (1979). Phylogenetic systematics (3rd edition of 1966 book). ISBN 0-252-06814-9.Translated from manuscript and so never published in German.
- Hull, David L. (1979). "The limits of cladism". Systematic Zoology 28: 416–440. doi:.
- Kitching, Ian J.; Peter L. Forey, Christopher J. Humphries and David M. Williams (1998). Cladistics: Theory and practice of parsimony analysis (2nd ed. ed.). Oxford University Press. ISBN 0-19-850138-2.
- Luria, Salvador; Stephen Jay Gould and Sam Singer (1981). A view of life. Menlo Park, CA: Benjamin/Cummings. ISBN 0-8053-6648-2.
- Mayr, Ernst (1982). The growth of biological thought: diversity, evolution and inheritance. Cambridge, MA: Harvard Univ. Press. ISBN 0-674-36446-5.
- Mayr, Ernst (1976). Evolution and the diversity of life (Selected essays). Cambridge, MA: Harvard Univ. Press. ISBN 0-674-27105-X. Reissued 1997 in paperback. Includes a reprint of Mayr's 1974 anti-cladistics paper at pp. 433-476, "Cladistic analysis or cladistic classification." This is the paper to which Hennig (1975) is a response.
- Patterson, Colin (1982). "Morphological characters and homology". Joysey, Kenneth A; A. E. Friday (editors) Problems in Phylogenetic Reconstruction, London: Academic Press.
- Rosen, Donn; Gareth Nelson and Colin Patterson (1979), Foreword provided for Hennig (1979)
- Shedlock, Andrew M; Norihiro Okada (2000). "SINE insertions: Powerful tools for molecular systematics". Bioessays 22: 148–160. doi:. ISSN 0039-7989. PMID 10655034.
- Sokal, Robert R. (1975). "Mayr on cladism -- and his critics". Systematic Zoology 24: 257–262. doi:.
- Swofford, David L.; G. J. Olsen, P. J. Waddell and David M. Hillis (1996). "Phylogenetic inference". in Hillis, David M; C. Moritz and B. K. Mable (editors). Molecular Systematics (2. ed. ed.). Sunderland, MA: Sinauer Associates. ISBN 0-87893-282-8.
- Wiley, Edward O. (1981). Phylogenetics: The Theory and Practice of Phylogenetic Systematics. New York: Wiley Interscience. ISBN 0-471-05975-7.
- Zwickl DJ, Hillis DM (2002). "Increased taxon sampling greatly reduces phylogenetic error". Systematic Biology 51: 588–598. doi:.
External links
- The Compleat Cladist (pdf)
- Tree of Life illustration - A high-level cladogram showing the complete tree of life.
- Example of cladistics used in textual criticism
- Journey into Phylogenetic Systematics
- Phylogenetics Primer from Talk.Origins
- Willi Hennig Society
- Cladistics: The International Journal of the Willi Hennig Society (ISSN 0748-3007)
- Phylogenetic inferring on the T-REX server
- A list of cladogram-generating programs
- For a cladistic approach to animal classification: Classification of living things
|
|||||||||||||||||