Clade
A clade is a taxonomic group comprising a single common ancestor and all the descendants of that ancestor.[1] Any such group is truly considered to be a monophyletic group,[note 1][2] and can be represented by both a phylogenetic analysis, as in a tree diagram, and by a cladogram (see cladistics), or simply as a taxonomic reference. "Clade" is derived from the ancient Greek κλάδος, klados, "branch."

If a clade proves robust in alternate cladistic analyses using different sets of data, it may be adopted into taxonomy and become a taxon. Not all taxa, however, are considered to be clades. Reptiles, for example, are a paraphyletic group because they do not include aves (birds), which are thought to also have evolved from the common ancestor of the reptiles.
In cladistics, a clade that is located within another more inclusive clade is said to be "nested" within that clade. Nested clade analysis is beneficial in many ways. For instance, it enables the detection of range expansions in isolated geographic areas.
Contents |
Relation to paraphyletic and polyphyletic groups
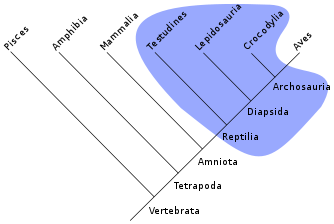

Groups that do not include all the descendants of the most recent common ancestor are said to be paraphyletic. For example, as shown in the adjacent illustration, reptiles are paraphyletic because that group excludes birds. A group that does not contain the most recent common ancestor of its members is said to be polyphyletic (Greek polys = many). An example of a polyphyletic group is the warm-blooded animals.
Because paraphyletic and polyphyletic groups are not monophyletic, they are not clades.
Naming clades
Valid taxa are monophyletic groups - that is, they are clades. However, traditional Linnaean taxonomy does not cope well in a cladistic framework: for instance, it cannot adequately classify stem groups.
Three methods of naming clades have been proposed: node-, stem-, and apomorphy-based. In node-based naming, taxon name A might refer to the least inclusive clade containing X and Y. In stem-based naming, A would refer to the most inclusive clade containing X and Y but not Z. In apomorphy (derived feature)-based naming, A would refer to the clade identified by a feature synapomorphic (sharing a derivation) with a feature in specimen (taxon) X. Differences between a traditional approach and these phylogenetic alternatives become obvious when the phylogenetic hypothesis changes.
Comparison between the traditional Linnaean approach to nomenclature and a phylogenetic alternative (node-based naming). Suppose that all we want to do is to give a name ("A") to a clade containing X and Y. In the Linnaean system this means that we have to introduce names for sister taxa, assigning all taxa to the categories species, genus, and family, and designate type species. No explicit reference to phylogeny is made. The phylogenetic alternative provides an explicit reference to evolutionary history, and nothing but the clade containing X and Y needs to be named. When the hypothesis of relationship changes, the phylogenetic alternative is cleaner and more explicit about what it refers to.
See also
- Phylogeny
- Paraphyly
- Polyphyly
- Phylogenetic nomenclature
- Binomial nomenclature
Notes
- ↑ The term 'monophyletic group' is used in this article in the conventional sense of 'an ancestor and all its descendants'. A case has been made that semantically, such groups should be referred to as 'holophyletic', but this term has not yet acquired widespread use. For more information, see Holophyletic
References
- ↑ "clade - Definitions from Dictionary.com". dictionary.reference.com. Retrieved on 2008-04-23.
- ↑ . doi:.
External links
- Evolving Thoughts: Clade
- A poster-sized cladogram showing the Tree of Life (Evolution)
- DM Hillis, D Zwickl & R Gutell: ~3000 species Tree of Life A cladogram?
- Phylogenetic systematics, an introductory slide-show on evolutionary trees University of California, Berkeley
|
|||||||||||||||||
|
|||||||||||||||||||||||||||||