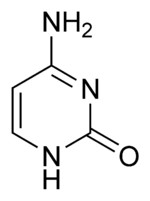
Cytosine
| Cytosine | |
|---|---|
 |
|
 |
 |
| IUPAC name | 4-amino-1H-pyrimidine-2-one |
| Identifiers | |
| CAS number | 71-30-7 |
| PubChem | |
| MeSH | |
| SMILES |
|
| Properties | |
| Molecular formula | C4H5N3O |
| Molar mass | 111.300 |
| Melting point |
320 - 325°C (decomp) |
| Except where noted otherwise, data are given for materials in their standard state (at 25 °C, 100 kPa) Infobox references |
|
Cytosine is one of the five main bases found in DNA and RNA. It is a pyrimidine derivative, with a heterocyclic aromatic ring and two substituents attached (an amine group at position 4 and a keto group at position 2). The nucleoside of cytosine is cytidine. In Watson-Crick base pairing, it forms three hydrogen bonds with guanine.
Contents |
History
Cytosine was discovered by Albrecht Kossel in 1894 when it was hydrolysed from calf thymus tissues.[1] A structure was proposed in 1903, and was synthesized (and thus confirmed) in the laboratory in the same year.
Cytosine recently found use in quantum computation. The first time any quantum mechanical properties were harnessed to process information took place on August 1st in 1998 when researchers at Oxford implemented David Deutsch's algorithm on a two qubit NMRQC (Nuclear Magnetic Resonance Quantum Computer) based on the cytosine molecule.[2]
Chemical reactions
Cytosine can be found as part of DNA, RNA, or as a part of a nucleotide. As cytidine triphosphate (CTP), it can act as a co-factor to enzymes, and can transfer a phosphate to convert adenosine diphosphate (ADP) to adenosine triphosphate (ATP).
In DNA and RNA, cytosine is paired with guanine. However, it is inherently unstable, and can change into uracil (spontaneous deamination). This can lead to a point mutation if not repaired by the DNA repair enzymes such as uracil glycosylase, which cleaves a uracil in DNA.
Cytosine can also be methylated into 5-methylcytosine by an enzyme called DNA methyltransferase.
References
- ↑ Kossel, A.; Steudel, H. Z. Physiol. Chem. 1903, 38, 49
- ↑ Jones, J.A.; M. Mosca (1998-08-01). "Implementation of a quantum algorithm on a nuclear magnetic resonance quantum computer". J.Chem.Phys 109 (109): 1648–1653. doi:. http://www.citebase.org/abstract?id=oai%3AarXiv.org%3Aquant-ph%2F9801027. Retrieved on 2007-10-18.
External links
- EINECS number 200-749-5
- Computational Chemistry Wiki
- Shapiro R (1999). "Prebiotic cytosine synthesis: a critical analysis and implications for the origin of life". Proc. Natl. Acad. Sci. U.S.A. 96 (8): 4396–401. PMID 10200273. http://www.pnas.org/cgi/content/full/96/8/4396.
|
|||||||||||||||||||||||||||