Race (classification of human beings)
2008/9 Schools Wikipedia Selection. Related subjects: Culture and Diversity
The term race or racial group usually refers to the concept of dividing humans into populations or groups on the basis of various sets of characteristics. The most widely used human racial categories are based on visible traits (especially skin colour, cranial or facial features and hair texture), and self-identification.
Conceptions of race, as well as specific ways of grouping races, vary by culture and over time, and are often controversial for scientific as well as social and political reasons. The controversy ultimately revolves around whether or not races are natural types or socially constructed, and the degree to which observed differences in ability and achievement, categorized on the basis of race, are a product of inherited (i.e. genetic) traits or environmental, social and cultural factors.
Some argue that although race is a valid taxonomic concept in other species, it cannot be applied to humans. Many scientists have argued that race definitions are imprecise, arbitrary, derived from custom, have many exceptions, have many gradations, and that the numbers of races delineated vary according to the culture making the racial distinctions; thus they reject the notion that any definition of race pertaining to humans can have taxonomic rigour and validity. Today most scientists study human genotypic and phenotypic variation using concepts such as "population" and " clinal gradation". Many contend that while racial categorizations may be marked by phenotypic or genotypic traits, the idea of race itself, and actual divisions of persons into races, are social constructs.
History
In ancient civilizations
Given visually complex social relationships, humans presumably have always observed and speculated about the physical differences among individuals and groups. But different societies have attributed markedly different meanings to these distinctions. For example, the Ancient Egyptian sacred text called Book of Gates identifies four categories that are now conventionally labeled "Egyptians", "Asiatics", "Libyans", and "Nubians", but such distinctions tended to conflate differences as defined by physical features such as skin tone, with tribal and national identity. Classical civilizations from Rome to China tended to invest much more importance in familial or tribal affiliation than with one's physical appearance (Dikötter 1992; Goldenberg 2003). Ancient Greek and Roman authors also attempted to explain and categorize visible biological differences among peoples known to them. Such categories often also included fantastical human-like beings that were supposed to exist in far-away lands. Some Roman writers adhered to an environmental determinism in which climate could affect the appearance and character of groups (Isaac 2004). In many ancient civilizations, individuals with widely varying physical appearances became full members of a society by growing up within that society or by adopting that society's cultural norms (Snowden 1983; Lewis 1990).
Julian the Apostate was an early observer of the differences in humans, based upon ethnic, cultural, and geographic traits, but as the ideology of "race" had not yet been constructed, he believed that they were the result of "Providence":
Come, tell me why it is that the Celts and the Germans are fierce, while the Hellenes and Romans are, generally speaking, inclined to political life and humane, though at the same time unyielding and warlike? Why the Egyptians are more intelligent and more given to crafts, and the Syrians unwarlike and effeminate, but at the same time intelligent, hot-tempered, vain and quick to learn? For if there is anyone who does not discern a reason for these differences among the nations, but rather declaims that all this so befell spontaneously, how, I ask, can he still believe that the universe is administered by a providence? — Julian, the Apostate.
Medieval models of "race" mixed Classical ideas with the notion that humanity as a whole was descended from Shem, Ham and Japheth, the three sons of Noah, producing distinct Semitic (Asian), Hamitic (African), and Japhetic (European) peoples.
Age of Discovery
The word "race", along with many of the ideas now associated with the term, were products of European imperialism and colonization during the age of exploration. (Smedley 1999) As Europeans encountered people from different parts of the world, they speculated about the physical, social, and cultural differences among various human groups. The rise of the Atlantic slave trade, which gradually displaced an earlier trade in slaves from throughout the world, created a further incentive to categorize human groups in order to justify the subordination of African slaves. (Meltzer 1993) Drawing on Classical sources and upon their own internal interactions — for example, the hostility between the English and Irish was a powerful influence on early thinking about the differences between people (Takaki 1993) — Europeans began to sort themselves and others into groups associated with physical appearance and with deeply ingrained behaviors and capacities. A set of folk beliefs took hold that linked inherited physical differences between groups to inherited intellectual, behavioural, and moral qualities. (Banton 1977) Although similar ideas can be found in other cultures (Lewis 1990; Dikötter 1992), they appear not to have had as much influence upon their social structures as was found in Europe and the parts of the world colonized by Europeans. However, often brutal conflicts between ethnic groups have existed throughout history and across the world.
Scientific concepts
The first scientific attempts to classify humans by categories of race date from the 17th century, along with the development of European imperialism and colonization around the world. The first post- Classical published classification of humans into distinct races seems to be François Bernier's Nouvelle division de la terre par les différents espèces ou races qui l'habitent ("New division of Earth by the different species or races which inhabit it"), published in 1684.
17th and 18th century
According to philosopher Michel Foucault, theories of both racial and class conflict can be traced to 17th century political debates about innate differences among ethnicities. In England radicals such as John Lilburne emphasised conflicts between Saxon and Norman peoples. In France Henri de Boulainvilliers argued that the Germanic Franks possessed a natural right to leadership, in contrast to descendants of the Gauls. In the 18th century, the differences among human groups became a focus of scientific investigation (Todorov 1993). Initially, scholars focused on cataloguing and describing " The Natural Varieties of Mankind," as Johann Friedrich Blumenbach entitled his 1775 text (which established the five major divisions of humans still reflected in some racial classifications, i.e., the Caucasoid race, Mongoloid race, Ethiopian race (later termed the Negroid race), American Indian race, and Malayan race). From the 17th through the 19th centuries, the merging of folk beliefs about group differences with scientific explanations of those differences produced what one scholar has called an " ideology of race" (Smedley 1999). According to this ideology, races are primordial, natural, enduring and distinct. It was further argued that some groups may be the result of mixture between formerly distinct populations, but that careful study could distinguish the ancestral races that had combined to produce admixed groups.
19th century
The 19th century saw attempts to change race from a taxonomic to a biological concept. In the 19th century a number of natural scientists wrote on race: Georges Cuvier, Charles Darwin, Alfred Wallace, Francis Galton, James Cowles Pritchard, Louis Agassiz, Charles Pickering, and Johann Friedrich Blumenbach. As the science of anthropology took shape in the 19th century, European and American scientists increasingly sought explanations for the behavioural and cultural differences they attributed to groups (Stanton 1960). For example, using anthropometrics, invented by Francis Galton and Alphonse Bertillon, they measured the shapes and sizes of skulls and related the results to group differences in intelligence or other attributes (Lieberman 2001).
These scientists made three claims about race: first, that races are objective, naturally occurring divisions of humanity; second, that there is a strong relationship between biological races and other human phenomena (such as forms of activity and interpersonal relations and culture, and by extension the relative material success of cultures), thus biologizing the notion of "race", as Foucault demonstrated in his historical analysis; third, that race is therefore a valid scientific category that can be used to explain and predict individual and group behaviour. Races were distinguished by skin colour, facial type, cranial profile and size, texture and colour of hair. Moreover, races were almost universally considered to reflect group differences in moral character and intelligence.
The eugenics movement of the late 19th and early 20th centuries, inspired by Arthur Gobineau's An Essay on the Inequality of the Human Races (1853–1855) and Vacher de Lapouge's "anthroposociology", asserted as self-evident the biological inferiority of particular groups (Kevles 1985). In many parts of the world, the idea of race became a way of rigidly dividing groups by culture as well as by physical appearances (Hannaford 1996). Campaigns of oppression and genocide were often motivated by supposed racial differences (Horowitz 2001).
In Charles Darwin's most controversial book, The Descent of Man, he made strong suggestions of racial differences and European superiority. In Darwin's view, stronger tribes of humans always replaced weaker tribes. As savage tribes came in conflict with civilized nations, such as England, the less advanced people were destroyed. Nevertheless, he also noted the great difficulty naturalists had in trying to decide how many "races" there actually were (Darwin was himself a monogenist on the question of race, believing that all humans were of the same species and finding "race" to be a somewhat arbitrary distinction among some groups):
Man has been studied more carefully than any other animal, and yet there is the greatest possible diversity amongst capable judges whether he should be classed as a single species or race, or as two (Virey), as three (Jacquinot), as four (Kant), five (Blumenbach), six (Buffon), seven (Hunter), eight (Agassiz), eleven (Pickering), fifteen (Bory St. Vincent), sixteen (Desmoulins), twenty-two (Morton), sixty (Crawfurd), or as sixty-three, according to Burke. This diversity of judgment does not prove that the races ought not to be ranked as species, but it shews that they graduate into each other, and that it is hardly possible to discover clear distinctive characters between them.
Modern debates
Models of human evolution
In a recent article, Leonard Lieberman and Fatimah Jackson have suggested that any new support for a biological concept of race will likely come from another source, namely, the study of human evolution. They therefore ask what, if any, implications current models of human evolution may have for any biological conception of race.
Today, all humans are classified as belonging to the species Homo sapiens and sub-species Homo sapiens sapiens. However, this is not the first species of hominids: the first species of genus Homo, Homo habilis, evolved in East Africa at least 2 million years ago, and members of this species populated different parts of Africa in a relatively short time. Homo erectus evolved more than 1.8 million years ago, and by 1.5 million years ago had spread throughout the Old World. Virtually all physical anthropologists agree that Homo sapiens evolved out of Homo erectus. Anthropologists have been divided as to whether Homo sapiens evolved as one interconnected species from H. erectus (called the Multiregional Model, or the Regional Continuity Model), or evolved only in East Africa, and then migrated out of Africa and replaced H. erectus populations throughout the Old World (called the Out of Africa Model or the Complete Replacement Model). Anthropologists continue to debate both possibilities, and the evidence is technically ambiguous as to which model is correct, although most anthropologists currently favour the Out of Africa model.
Multiregional model
Advocates of the Multiregional model, primarily Milford Wolpoff and his associates, have argued that the simultaneous evolution of H. sapiens in different parts of Europe and Asia would have been possible if there were a degree of gene flow between archaic populations. Similarities of morphological features between archaic European and Chinese populations and modern H. sapiens from the same regions, Wolpoff argues, support a regional continuity only possible within the Multiregional model. Wolpoff and others further argue that this model is consistent with clinal patterns of phenotypic variation (Wolpoff 1993). Lieberman and Jackson have related this theory to race with the following statement:
| “ | The major implication for race in the multiregional evolution continuity model involves the time depth of a million or more years in which race differentiation might evolve in diverse ecological regions [...]. This must be balanced against the degree of gene flow and the transregional operation of natural selection on encephalization due to development of tools and, more broadly, culture. | ” |
Out of Africa model
According to the Out of Africa Model, developed by Christopher Stringer and Peter Andrews, modern Homo sapiens evolved in Africa 200,000 years ago. H. sapiens began migrating from Africa around 50,000 years ago and eventually replaced existing hominid species in Europe and Asia. This model has gained support by recent research using mitochondrial DNA (mtDNA). After analysing genealogy trees constructed using 133 types of mtDNA, they concluded that all were descended from a woman from Africa, dubbed Mitochondrial Eve. Lieberman and Jackson have related this theory to race with the following comment:
| “ | There are three major implications of this model for the race concept. First, the shallow time dimension minimizes the degree to which racial differences could have evolved [...]. Second, the mitochondrial DNA model presents a view that is very much different from Carleton Coon's (1962) concerning the time at which Africans passed the threshold from archaic to modern, thereby minimizing race differences and avoiding racist implications. However, the model, as interpreted by Wainscoat et al. (1989:34), does describe "a major division of human populations into an African and a Eurasian group." This conclusion could best be used to emphasize the degree of biological differences, and thereby provide support for the race concept. Third, the replacement of preexisting members of genus Homo (with little gene flow) implies several possible causes from disease epidemics to extermination. If the latter, then from a contemporary viewpoint, xenophobia or racism may have been practiced" | ” |
Comparison of the two models
Lieberman and Jackson have argued that while advocates of both the Multiregional Model and the Out of Africa Model use the word race and make racial assumptions, none define the term. They conclude that "Each model has implications that both magnify and minimize the differences between races. Yet each model seems to take race and races as a conceptual reality. The net result is that those anthropologists who prefer to view races as a reality are encouraged to do so" and conclude that students of human evolution would be better off avoiding the word race, and instead describe genetic differences in terms of populations and clinal gradations.
Race as subspecies
With the advent of the modern synthesis in the early 20th century, many biologists sought to use evolutionary models and populations genetics in an attempt to formalise taxonomy below the species level. The term subspecies is used by biologists when a group of organisms are classified in such a way. In biology the term "race" is very rarely used because it is ambiguous, "'Race' is not being defined or used consistently; its referents are varied and shift depending on context. The term is often used colloquially to refer to a range of human groupings. Religious, cultural, social, national, ethnic, linguistic, genetic, geographical and anatomical groups have been and sometimes still are called 'races'". Generally when it is used it is synonymous with subspecies. One of the main obstacles to identifying subspecies is that, while it is a recognised taxonomic term, it has no precise definition.
Some species of organisms do not appear to fragment into subgroups, while others do seem to form such subspecific groups. A monotypic species comprises a single group or rather a single subspecies. Monotypic species can occur in several ways:
- All members of the species are very similar and cannot be sensibly divided into biologically significant subcategories.
- The individuals vary considerably but the variation is essentially random and largely meaningless so far as genetic transmission of these variations is concerned (many plant species fit into this category, which is why horticulturists interested in preserving, say, a particular flower colour avoid propagation from seed, and instead use vegetative methods like propagation from cuttings).
- The variation among individuals is noticeable and follows a pattern, but there are no clear dividing lines among separate groups: they fade imperceptibly into one another. Such clinal variation always indicates substantial gene flow among the apparently separate groups that make up the population(s). Populations that have a steady, substantial gene flow among them are likely to represent a monotypic species even when a fair degree of genetic variation is obvious.
A polytypic species has two or more subspecies. These are separate populations that are more genetically different from one another and that are more reproductively isolated, gene flow between these populations is much reduced leading to genetic differentiation.
Morphological subspecies
Traditionally subspecies are seen as geographically isolated and genetically differentiated populations. Or to put it another way "the designation 'subspecies' is used to indicate an objective degree of microevolutionary divergence" One objection to this idea is that it does not identify any degree of differentiation, therefore any population that is somewhat biologically different could be considered a subspecies, even to the level of a local population. As a result it is necessary to impose a threshold on the level of difference that is required for a population to be designated a subspecies. This effectively means that populations of organisms must have reached a certain measurable level of difference in order to be recognised as subspecies. Dean Amadon proposed in 1949 that subspecies would be defined according to the seventy-five percent rule which means that 75% of a population must lie outside 99% of the range of other populations for a given defining morphological character or a set of characters. The 75 percent rule still has defenders but other scholars argue that it should be replaced with 90 or 95 percent rule.
When biologists study non-human populations, the standard threshold at which morphological diversity between two different populations is considered differentiated enough to be classified as subspecies is set at 70-75%. Smith et al. write:
The non-discrete nature of subspecies is evident from their definition as geographic segments of any given gonochoristic (bisexually reproducing) species differing from each other to a reasonably practical degree (e.g., at least 70-75%), but to less than totality. All subspecies are allopatric (either dichopatric [with non-contiguous ranges] or parapatric [with contiguous ranges], except for cases of circular overlap with sympatry); sympatry is conclusive evidence (except for cases of circular overlap) of allospecificity (separate specific status). Parapatric subspecies interbreed and exhibit intergradation in contact zones, but such taxa maintain the required level of distinction in one or more characters outside of those zones. Dichopatric populations are regarded as subspecies if they fail to exhibit full differentiation (i.e., exhibit overlap in variation of their differentiae up to 25-30%), even in the absence of contact (overlap exceeding 25-30% does not qualify for taxonomic recognition of either dichopatric populations or of parapatric populations outside of their zones of intergradation). Phenotypic adjustment to differing environmental conditions through natural selection is likely the primary factor in divergence of parapatric subspecies, and undoubtedly is involved in some dichopaffic subspecies. The founder effect and genetic drift are involved more in the latter than in the former.
Thus, according to the seventy-five percent rule two populations represent different subspecies if the morphological differences between them reach between 25-30%.
In 1978, Sewall Wright suggested that human populations that have long inhabited separated parts of the world should, in general, be considered to be of different subspecies by the usual criterion that most individuals of such populations can be allocated correctly by inspection. It does not require a trained anthropologist to classify an array of Englishmen, West Africans, and Chinese with 100% accuracy by features, skin colour, and type of hair in spite of so much variability within each of these groups that every individual can easily be distinguished from every other. However, it is customary to use the term race rather than subspecies for the major subdivisions of the human species as well as for minor ones.
Humans can be correctly assigned to races at much greater than 75% accuracy on the basis of morphological traits while chimpanzee subspecies are morphologically indistinct, and difficult or impossible to classify when raised in captivity.
On the other hand in practice subspecies are often defined by easily observable physical appearance, but there is not necessarily any evolutionary significance to these observed differences, so this form of classification is generally not accepted by evolutionary biologists.
Because of the difficulty in classifying subspecies morphologically, many biologists began to reject the concept altogether, citing problems such as:
- Visible physical differences do not correlate with one another, leading to the possibility of different classifications for the same individual organisms.
- Parallel evolution can lead to the existence of the appearance of similarities between groups of organisms that are not part of the same species.
- The existence of isolated populations within previously designated subspecies.
- That the criteria for classification are arbitrary.
Subspecies as isolated differentiated populations
Genetic differences between populations of organisms can be determined using the fixation index of Sewall Wright, which is often abbreviated to FST. This statistic is used to compare differences between any two given populations. For example it is often stated that the fixation index for humans is about 0.15. This means that about 85% of the variation measured in the human population is within any population, and about 15% of the variation occurs between populations.
Citing Smith, Templeton asserts that an FST value of 0.25 or 0.30 between populations is a “standard criterion” for subspecies classification. However, Smith doesn't mention the concept of FST in his paper, he is discussing variation in morphology and discusses the 75% rule. Templeton reported that the white-tailed deer has an FST of about 60% and the grey wolf has an FST approaching 90% for mtDNA. The FST of grey wolves is 0.168 in autosomal loci, however. On the other hand, in a paper reporting on the phylogenetic structure of the Leopard Panthera pardus species of Africa and Asia, Uphyrkina et al. found that 76.04% of mtDNA variation was distributed between leopard populations and 23.96% within populations, for microsatellite autosomal data, 0.358 (35.8%) of the variation was found between populations. This compares to an mtDNA FST of between 0.24-0.27 (24-27%), and a genomic FST of about 0.15 (15%) for humans, and an FST of 0.09-0.32 for autosomal microsatellite DNA between three Chimpanzee (Pan troglodytes) populations and of 0.51-0.68 between these three populations and the bonobo (Pan paniscus) populations. The following guidelines were suggested by Sewall Wright for interpreting FST:
“The range 0 to 0.05 may be considered as indicating little genetic differentiation. The range 0.05 to 0.15 indicates moderate genetic differentiation. The range 0.15 to 0.25 indicates great genetic differentiation. Values of FST above 0.25 indicate very great genetic differentiation.”
Wright found differences in FST for various species from 0.023-0.501. For humans the FST is usually given as 0.15, of this 15% that is distributed between populations about 3-6% is distributed between geographically close populations occupying the same continent and about 6-10% is distributed between more distant continental groups, these figures vary somewhat depending on the type of genetic systems used, but the general observation has been reproduced in numerous studies. This indicates that some of the between population variation for humans is found within any "race" and about 6-10% of variation is found between "races", giving an FST of 0.06-0.1 for human "races".
It has also been noted that:
- "First, compared with many other mammalian species, humans are genetically less diverse... For example, the chimpanzee subspecies living just in central and western Africa have higher levels of diversity than do humans (Ebersberger et al. 2002; Yu et al. 2003; Fischer et al. 2004)." Human variation is also distributed in an unusual and not easily understood fashion compared to other mammalian species: "The details of this distribution are impossible to describe succinctly because of the difficulty of defining a 'population,' the clinal nature of variation, and heterogeneity across the genome (Long and Kittles 2003).... This distribution of genetic variation differs from the pattern seen in many other mammalian species, for which existing data suggest greater differentiation between groups (Templeton 1998; Kittles and Weiss 2003)."
- "Since the 1980s, there have been indications that the genetic diversity of humans is low compared with that of many other species. This has been interpreted to mean that humans are a relatively young species, so populations have had relatively little time to differentiate from one another. For example, 2 randomly chosen humans differ at ~1 in 1,000 nucleotide pairs, whereas two chimpanzees differ at ~1 in 500 nucleotide pairs.
- "'Race' is a legitimate taxonomic concept that works for chimpanzees but does not apply to humans (at this time). The nonexistence of 'races' or subspecies in modern humans does not preclude substantial genetic variation that may be localized to regions or populations....The DNA of an unknown individual from one of the sampled populations would probably be correctly linked to a population. Because this identification is possible does not mean that there is a level of differentiation equal to 'races'. The genetics of Homo sapiens shows gradients of differentiation."
- "Humans are ~98.8% similar to chimpanzees at the nucleotide level and are considerably more similar to each other, differing on average at only 1 of every 500−1,000 nucleotides between chromosomes. This degree of diversity is less than what typically exists among chimpanzees.
- "The average proportion of nucleotide differences between a randomly chosen pair of humans (i.e., average nucleotide diversity, or π) is consistently estimated to lie between 1 in 1,000 and 1 in 1,500. This proportion is low compared with those of many other species, from fruit flies to chimpanzees...."
Population genetics: population and cline
At the beginning of the 20th century, anthropologists questioned, and eventually abandoned, the claim that biologically distinct races are isomorphic with distinct linguistic, cultural, and social groups. Shortly thereafter, the rise of population genetics provided scientists with a new understanding of the sources of phenotypic variation. This new science has led many mainstream evolutionary scientists in anthropology and biology to question the very validity of race as a scientific concept describing an objectively real phenomenon. Those who came to reject the validity of the concept of race did so for four reasons: empirical, definitional, the availability of alternative concepts, and ethical (Lieberman and Byrne 1993).
The first to challenge the concept of race on empirical grounds were anthropologists Franz Boas, who demonstrated phenotypic plasticity due to environmental factors (Boas 1912), and Ashley Montagu (1941, 1942), who relied on evidence from genetics. Zoologists Edward O. Wilson and W. Brown then challenged the concept from the perspective of general animal systematics, and further rejected the claim that "races" were equivalent to "subspecies" (Wilson and Brown 1953).
Clines
One of the crucial innovations in reconceptualizing genotypic and phenotypic variation was anthropologist C. Loring Brace's observation that such variations, insofar as it is affected by natural selection, migration, or genetic drift, are distributed along geographic gradations or clines (Brace 1964). This point called attention to a problem common to phenotype-based descriptions of races (for example, those based on hair texture and skin colour): they ignore a host of other similarities and differences (for example, blood type) that do not correlate highly with the markers for race. Thus, anthropologist Frank Livingstone's conclusion that, since clines cross racial boundaries, "there are no races, only clines" (Livingstone 1962: 279).
In a response to Livingston, Theodore Dobzhansky argued that when talking about "race" one must be attentive to how the term is being used: "I agree with Dr. Livingston that if races have to be 'discrete units,' then there are no races, and if 'race' is used as an 'explanation' of the human variability, rather than vice versa, then the explanation is invalid." He further argued that one could use the term race if one distinguished between "race differences" and "the race concept." The former refers to any distinction in gene frequencies between populations; the latter is "a matter of judgment." He further observed that even when there is clinal variation, "Race differences are objectively ascertainable biological phenomena .... but it does not follow that racially distinct populations must be given racial (or subspecific) labels." In short, Livingston and Dobzhansky agree that there are genetic differences among human beings; they also agree that the use of the race concept to classify people, and how the race concept is used, is a matter of social convention. They differ on whether the race concept remains a meaningful and useful social convention.
In 1964, biologists Paul Ehrlich and Holm pointed out cases where two or more clines are distributed discordantly—for example, melanin is distributed in a decreasing pattern from the equator north and south; frequencies for the haplotype for beta-S hemoglobin, on the other hand, radiate out of specific geographical points in Africa (Ehrlich and Holm 1964). As anthropologists Leonard Lieberman and Fatimah Linda Jackson observe, "Discordant patterns of heterogeneity falsify any description of a population as if it were genotypically or even phenotypically homogeneous" (Lieverman and Jackson 1995).
Patterns such as those seen in human physical and genetic variation as described above, have led to the consequence that the number and geographic location of any described races is highly dependent on the importance attributed to, and quantity of, the traits considered. For example if only skin colour and a "two race" system of classification were used, then one might classify Indigenous Australians in the same "race" as Black people, and Caucasians in the same "race" as East Asian people, but biologists and anthropologists would dispute that these classifications have any scientific validity. On the other hand the greater the number of traits (or alleles) considered, the more subdivisions of humanity are detected, due to the fact that traits and gene frequencies do not always correspond to the same geographical location, or as Ossario and Duster (2005) put it:
Anthropologists long ago discovered that humans' physical traits vary gradually, with groups that are close geographic neighbors being more similar than groups that are geographically separated. This pattern of variation, known as clinal variation, is also observed for many alleles that vary from one human group to another. Another observation is that traits or alleles that vary from one group to another do not vary at the same rate. This pattern is referred to as nonconcordant variation. Because the variation of physical traits is clinal and nonconcordant, anthropologists of the late 19th and early 20th centuries discovered that the more traits and the more human groups they measured, the fewer discrete differences they observed among races and the more categories they had to create to classify human beings. The number of races observed expanded to the 30s and 50s, and eventually anthropologists concluded that there were no discrete races (Marks, 2002). Twentieth and 21st century biomedical researchers have discovered this same feature when evaluating human variation at the level of alleles and allele frequencies. Nature has not created four or five distinct, nonoverlapping genetic groups of people.
Populations
Population geneticists have debated as to whether the concept of population can provide a basis for a new conception of race. In order to do this a working definition of population must be found. Surprisingly there is no generally accepted concept of population that biologists use. It has been pointed out that the concept of population is central to ecology, evolutionary biology and conservation biology, but also that most definitions of population rely on qualitative descriptions such as "a group of organisms of the same species occupying a particular space at a particular time" Waples and Gaggiotti identify two broad types of definitions for populations, those that fall into an ecological paradigm and those that fall into an evolutionary paradigm. Examples such definitions are:
- Ecological paradigm: A group of individuals of the same species that co-occur in space and time and have an opportunity to interact with each other.
- Evolutionary paradigm: A group of individuals of the same species living in close enough proximity that any member of the group can potentially mate with any other member.
Richard Lewontin, claiming that 85 percent of human variation occurs within populations, and not among populations, argued that neither "race" nor "subspecies" were appropriate or useful ways to describe populations (Lewontin 1973). Nevertheless, barriers—which may be cultural or physical— between populations can limit gene flow and increase genetic differences. Recent work by population geneticists conducting research in Europe suggests that ethnic identity can be a barrier to gene flow. Others, such as Ernst Mayr, have argued for a notion of "geographic race" . Some researchers report the variation between racial groups (measured by Sewall Wright's population structure statistic FST) accounts for as little as 5% of human genetic variation². Sewall Wright himself commented that if differences this large were seen in another species, they would be called subspecies. In 2003 A. W. F. Edwards argued that cluster analysis supersedes Lewontin's arguments (see below).
These empirical challenges to the concept of race forced evolutionary sciences to reconsider their definition of race. Mid-century, anthropologist William Boyd defined race as:
- A population which differs significantly from other populations in regard to the frequency of one or more of the genes it possesses. It is an arbitrary matter which, and how many, gene loci we choose to consider as a significant "constellation" (Boyd 1950).
Lieberman and Jackson (1994) have pointed out that "the weakness of this statement is that if one gene can distinguish races then the number of races is as numerous as the number of human couples reproducing." Moreover, anthropologist Stephen Molnar has suggested that the discordance of clines inevitably results in a multiplication of races that renders the concept itself useless (Molnar 1992).
The distribution of many physical traits resembles the distribution of genetic variation within and between human populations (American Association of Physical Anthropologists 1996; Keita and Kittles 1997). For example, ~90% of the variation in human head shapes occurs within every human group, and ~10% separates groups, with a greater variability of head shape among individuals with recent African ancestors (Relethford 2002).
Molecular genetics: lineages and clusters
With the recent availability of large amounts of human genetic data from many geographically distant human groups scientists have again started to investigate the relationships between people from various parts of the world. One method is to investigate DNA molecules that are passed down from mother to child (mtDNA) or from father to son (Y chromosomes), these form molecular lineages and can be informative regarding prehistoric population migrations. Alternatively autosomal alleles are investigated in an attempt to understand how much genetic material groups of people share. This work has led to a debate amongst geneticists, molecular anthropologists and medical doctors as to the validity of conceps such as "race". Some researchers insist that classifying people into groups based on ancestry may be important from medical and social policy points of view, and claim to be able to do so accurately. Others claim that individuals from different groups share far too much of their genetic material for group membership to have any medical implications. This has reignited the scientific debate over the validity of human classification and concepts of "race".
Molecular lineages, Y chromosomes and mitochondrial DNA
Mitochondria are small organelles that lie in the cytoplasm of eucaryotic cells, such as those of humans. Their primary purpose is to provide energy to the cell. Mitochondria are thought to be the vestigial remains of symbiotic bacteria that were once free living. One indication that mitochondria were once free living is that they contain a relatively small circular segment of DNA, called mitochondrial DNA (mtDNA). The overwhelming majority of a human's DNA is contained in chromosomes in the nucleus of the cell, but mtDNA is an exception. An individual inherits their cytoplasm and the organelles it contains exclusively from their mother, as these are derived from the ovum (egg cell), sperm only carry chromosomal DNA due to the necessity of maintaining motility. When a mutation arises in mtDNA molecule the mutation is therefore passed in a direct female line of descent. These mutations are derived from copying mistakes, when the DNA is copied it is possible that a single mistake occurs in the DNA sequence, these single mistakes are called single nucleotide polymorphisms (SNPs).
Human Y chromosomes are male specific sex chromosomes, any human that possesses a Y chromosome will be morphologically male. Y chromosomes are therefore passed from father to son, although Y chromosomes are situated in the cell nucleus, they only recombine with the X chromosome at the ends of the Y chromosome, the vast majority of the Y chromosome (95%) does not recombine. Therefore, as with mtDNA, when mutations (SNPs) arise in the Y chromosome they are passed on directly from father to son in a direct male line of descent.
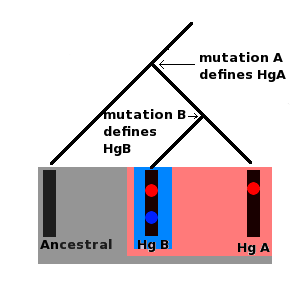
The Y chromosome and mtDNA therefore share certain properties. Other chromosomes, autosomes and X chromosomes in women, share their genetic material (called crossing over leading to recombination) during meiosis (a special type of cell division that occurs for the purposes of sexual reproduction). Effectively this means that the genetic material from these chromosomes gets mixed up in every generation, and so any new mutations are passed down randomly from parents to offspring. The special feature that both Y chromosomes and mtDNA display is that mutations can accrue along a certain segment of both molecules and these mutations remain fixed in place on the DNA. Furthermore the historical sequence of these mutations can also be inferred. For example, if a set of ten Y chromosomes (derived from ten different men) contains a mutation, A, but only five of these chromosomes contain a second mutation, B, it must be the case that mutation B occurred after mutation A. Furthermore all ten men who carry the chromosome with mutation A are the direct male line descendants of the same man who was the first person to carry this mutation. The first man to carry mutation B was also a direct male line descendant of this man, but is also the direct male line ancestor of all men carrying mutation B. Series of mutations such as this form molecular lineages. Furthermore each mutation defines a set of specific Y chromosomes called a haplogroup. All men carrying mutation A form a single haplogroup, all men carrying mutation B are part of this haplogroup, but mutation B also defines a more recent haplogroup (which is a subgroup or subclade) of its own which men carrying only mutation A do not belong to. Both mtDNA and Y chromosomes are grouped into lineages and haplogroups, these are often presented as tree like diagrams.
Groundbreaking work by molecular biologists such as Cann et al. (1987) on mtDNA produced three interesting observations relevant to race and human evolution.
Firstly, by estimating the rate at which mutations occur in mtDNA Cann et al. were able to estimate the age of the common ancestral mtDNA type: "the common ancestral mtDNA (type a) links mtDNA types that have diverged by an average of nearly 0.57%. Assuming a rate of 2%-4% per million years, this implies that the common ancestor of all surviving mtDNA types existed 140,000-290,000 years ago." This observation is robust, and this common direct female line ancestor (or mitochondrial most recent common ancestor (mtMRCA)) of all extant humans has become known as mitochondrial eve. The observation that the mtMRCA is the direct matrilineal ancestor of all living humans should not be interpreted as meaning that either she was the first anatomically modern human, nor that there were no other female humans living concurrently with her. A more reasonable explanation is that other women who lived at the same time as mtMRCA did indeed reproduce and pass their genes down to living humans, but that their mitochondrial lineages have been lost over time, probably due to random events such as producing only male children. It is impossible to know to what extent these non-extant lineages have been lost or how much they differed from the mtDNA of our mtMRCA. Cann et al.
Secondly Cann et al. postulate that their work supports an African origin for modern human mtDNA: "We infer from the tree of minimum length... that Africa is a likely source of the human mitochondrial gene pool. This inference comes from the observation that one of the two primary branches leads exclusively to African mtDNAs... while the second primary branch also leads to African mtDNAs... By postulating that the common ancestral mtDNA (type a in Fig. 3) was African, we minimize the number of intercontinental migrations needed to account for the geographic distribution of mtDNA types."
Thirdly the study shows that mtDNA types (haplogroups) do not cluster by geography, ethnicity or race, implying multiple female lineages were involved in founding modern human populations, with many closely related lineages spread geographically and many populations containing distantly related lineages: "The second implication of the tree (Fig. 3) -that each non-African population has multiple origins-can be illustrated most simply with the New Guineans. Take, as an example, mtDNA type 49, a lineage whose nearest relative is not in New Guinea, but in Asia (type 50). Asian lineage 50 is closer genealogically to this New Guinea lineage than to other Asian mtDNA lineages. Six other lineages lead exclusively to New Guinean mtDNAs, each originating at a different place in the tree (types 12, 13, 26-29, 65, 95 and 127-134 in Fig. 3). This small region of New Guinea (mainly the Eastern Highlands Province) thus seems to have been colonised by at least seven maternal lineages (Tables 2 and 3). In the same way, we calculate the minimum numbers of female lineages that colonised Australia, Asia and Europe (Tables 2 and 3). Each estimate is based on the number of region-specific clusters in the tree (Fig. 3, Tables 2 and 3). These numbers, ranging from 15 to 36 (Tables 2 and 3), will probably rise as more types of human mtDNA are discovered."
The Y chromosome is much larger than mtDNA, and is relatively homogeneous, therefore it has taken much longer to find distinct lineages and to analyse them. Conversely, because the Y chromosome is so large by comparison it can hold a great deal more genetic information. With regard to the three observations made by Cann et al. concerning mtDNA, Y chromosome studies show similar patterns. The estimate for the age of the ancestral Y chromosome for all extant Y chromosomes is given at about 70,000 years ago and is also placed in Africa, this individual is sometimes referred to as Y chromosome Adam. The difference in dates between Y chromosome Adam and mitochondrial Eve is usually attributed to a higher extinction rate for Y chromosomes due to greater differential reproductive success between individual men, that means that a small number of very successful men may produce a great many children, while a larger number of less successful men will produce far less children. Keita et al. (2004) say, with reference to Y chromosome and mtDNA and concepts of race:
Y-chromosome and mitochondrial DNA genealogies are especially interesting because they demonstrate the lack of concordance of lineages with morphology and facilitate a phylogenetic analysis. Individuals with the same morphology do not necessarily cluster with each other by lineage, and a given lineage does not include only individuals with the same trait complex (or 'racial type'). Y-chromosome DNA from Africa alone suffices to make this point. Africa contains populations whose members have a range of external phenotypes. This variation has usually been described in terms of 'race' (Caucasoids, Pygmoids, Congoids, Khoisanoids). But the Y-chromosome clade defined by the PN2 transition (PN2/M35, PN2/M2) [see haplogroup E3b and Haplogroup E3a] shatters the boundaries of phenotypically defined races and true breeding populations across a great geographical expanse21. African peoples with a range of skin colors, hair forms and physiognomies have substantial percentages of males whose Y chromosomes form closely related clades with each other, but not with others who are phenotypically similar. The individuals in the morphologically or geographically defined 'races' are not characterized by 'private' distinct lineages restricted to each of them.
How much are genes shared? Clustering analyses and what they tell us
Genetic data can be used to infer population structure and assign individuals to groups that often correspond with their self-identified geographical ancestry. Recently, Lynn Jorde and Steven Wooding argued that "Analysis of many loci now yields reasonably accurate estimates of genetic similarity among individuals, rather than populations. Clustering of individuals is correlated with geographic origin or ancestry."
In 2003 A. W. F. Edwards wrote a paper called Lewontin's Fallacy, rebuking the argument that because most of the variation is within group classification of humans is not possible. He claimed that this conclusion ignores the fact that most of the information that distinguishes populations is hidden in the correlation structure of the data and not simply in the variation of the individual factors. Edwards concludes that "It is not true that 'racial classification is ... of virtually no genetic or taxonomic significance' or that 'you can't predict someone’s race by their genes'." Likewise Neil Risch of Stanford University has proposed that self-identified race/ethnic group could be a valid means of categorization in the USA for public health and policy considerations. While a 2002 paper by Noah Rosenberg's group makes a similar claim "The structure of human populations is relevant in various epidemiological contexts. As a result of variation in frequencies of both genetic and nongenetic risk factors, rates of disease and of such phenotypes as adverse drug response vary across populations. Further, information about a patient’s population of origin might provide health care practitioners with information about risk when direct causes of disease are unknown."
Researchers such as Neil Risch and Noah Rosenberg have argued that a person's biological and cultural background may have important implications for medical treatment decisions, for example an opinion paper by Neil Risch's group in 2002 states:
Both for genetic and non-genetic reasons, we believe that racial and ethnic groups should not be assumed to be equivalent, either in terms of disease risk or drug response.....Whether African Americans, Hispanics, Native Americans, Pacific Islanders or Asians respond equally to a particular drug is an empirical question that can only be addressed by studying these groups individually.
While another 2002 paper by Noah Rosenberg's group makes a similar claim
The structure of human populations is relevant in various epidemiological contexts. As a result of variation in frequencies of both genetic and nongenetic risk factors, rates of disease and of such phenotypes as adverse drug response vary across populations. Further, information about a patient’s population of origin might provide health care practitioners with information about risk when direct causes of disease are unknown.
This work used samples from the Human Genome Diversity Project (HGDP), a project that has collected samples from individuals from 52 ethnic groups from various locations around the world. The HGDP has itself been criticised for collecting samples on an "ethnic group" basis, on the grounds that ethnic groups represent constructed categories rather than categories which are solely natural or biological. The molecular anthropologist Jonathan Marks states:
As any anthropologist knows, ethnic groups are categories of human invention, not given by nature. Their boundaries are porous, their existence historically ephemeral. There are the French, but no more Franks; there are the English, but no Saxons; and Navajos, but no Anasazi...we cannot really know the nature of the actual relationship of the modern group to the ancient one...The worst mistake you can make in human biology is to confuse constructed categories with natural ones. And to overload a big project with cultural categories as the overall sampling strategy would be a serious problem
In the same issue of Science that published the Rosenberg data, Mary-Claire King and Arno G. Motulsky give a similar warning regarding the HGDP data:
The identification of clusters corresponding to the major geographic regions may depend on the sampling of individuals from well-defined, relatively homogeneous populations. If individuals were sampled from a worldwide 'grid' (or a worldwide grid weighted by population density), the clusters might be much less precisely defined. Does the correspondence of worldwide genetic clusters and major geographic regions suggest borders around genetic clusters analogous to the physical borders—oceans, mountain ranges, and deserts—separating geographic regions? No. Both the results of Rosenberg and colleagues and those of previous studies indicate that unlike separations between geographic regions, differences in allele frequencies are gradual.
Another study by Neil Risch in 2005 used 326 microsatellite markers and self-identified race/ethnic group (SIRE), white (European American), African-American (black), Asian and Hispanic (individuals involved in the study had to choose from one of these categories), to representing discrete "populations", and showed distinct and non-overlapping clustering of the white, African-American and Asian samples. The results were claimed to confirm the integrity of self-described ancestry: "We have shown a nearly perfect correspondence between genetic cluster and SIRE for major ethnic groups living in the United States, with a discrepancy rate of only 0.14%." But also warned that: "This observation does not eliminate the potential for confounding in these populations. First, there may be subgroups within the larger population group that are too small to detect by cluster analysis. Second, there may not be discrete subgrouping but continuous ancestral variation that could lead to stratification bias. For example, African Americans have a continuous range of European ancestry that would not be detected by cluster analysis but could strongly confound genetic case-control studies. (Tang, 2005)
Studies such as those by Risch and Rosenberg use a computer program called STRUCTURE to find human populations (gene clusters). It is a statistical program that works by placing individuals into one of two clusters based on their overall genetic similarity, many possible pairs of clusters are tested per individual to generate multiple clusters. These populations are based on multiple genetic markers that are often shared between different human populations even over large geographic ranges. The notion of a genetic cluster is that people within the cluster share on average similar allele frequencies to each other than to those in other clusters. (Edwards, 2003 but see also infobox "Multi Locus Allele Clusters") In a test of idealised populations, the computer programme STRUCTURE was found to consistently under-estimate the numbers of populations in the data set when high migration rates between populations and slow mutation rates (such as single nucleotide polymorphisms) were considered.
Nevertheless the Rosenberg et al. (2002) paper shows that individuals can be assigned to specific clusters to a high degree of accuracy. One of the underlying questions regarding the distribution of human genetic diversity is related to the degree to which genes are shared between the observed clusters. It has been observed repeatedly that the majority of variation observed in the global human population is found within populations. This variation is usually calculated using Sewall Wright's Fixation index (FST), which is an estimate of between to within group variation. The degree of human genetic variation is a little different depending upon the gene type studied, but in general it is common to claim that ~85% of genetic variation is found within groups, ~6-10% between groups within the same continent and ~6-10% is found between continental groups. For example The Human Genome Project states "two random individuals from any one group are almost as different [genetically] as any two random individuals from the entire world." On the other hand Edwards (2003) claims in his essay " Lewontin's Fallacy" that: "It is not true, as Nature claimed, that 'two random individuals from any one group are almost as different as any two random individuals from the entire world'" and Risch et al. (2002) state "Two Caucasians are more similar to each other genetically than a Caucasian and an Asian." It should be noted that these statements are not the same. Risch et al. simply state that two indigenous individuals from the same geographical region are more similar to each other than either is to an indigenous individual from a different geographical region, a claim few would argue with. Jorde et al put it like this:
The picture that begins to emerge from this and other analyses of human genetic variation is that variation tends to be geographically structured, such that most individuals from the same geographic region will be more similar to one another than to individuals from a distant region.
Whereas Edwards claims that it is not true that the differences between individuals from different geographical regions represent only a small proportion of the variation within the human population (he claims that within group differences between individuals are not almost as large as between group differences). Bamshad et al. (2004) used the data from Rosenberg et al. (2002) to investigate the extent of genetic differences between individuals within continental groups relative to genetic differences between individuals between continental groups. They found that though these individuals could be classified very accurately to continental clusters, there was a significant degree of genetic overlap on the individual level, to the extent that, using 377 loci, individual Europeans were about 38% of the time more genetically similar to East Asians than to other Europeans.
The results obtained by clustering analyses are dependent on several criteria:
- The clusters produced are relative clusters and not absolute clusters, each cluster is the product of comparisons between sets of data derived for the study, results are therefore highly influenced by sampling strategies. (Edwards, 2003)
- The geographic distribution of the populations sampled, because human genetic diversity is marked by isolation by distance, populations from geographically distant regions will form much more discrete clusters than those from geographically close regions. (Kittles and Weiss, 2003)
- The number of genes used. The more genes used in a study the greater the resolution produced and therefore the greater number of clusters that will be identified. (Tang, 2005)
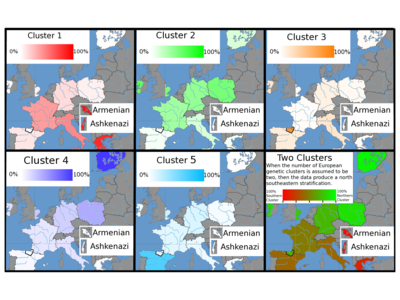
Additionally two studies of European population clusters have been produced. Seldin et al. (2006) identified three European clusters using 5,700 genome-wide polymorphisms. Bauchet et al. (2007) used 10,000 polymorphisms to identify five distinct clusters in the European population, consisting of a south-eastern European cluster (including samples from southern Italians, Armenian, Ashkenazi Jewish and Greek "populations"); a northern-European Cluster (including samples from German, eastern English, Polish and western Irish "populations"); a Basque cluster (including samples from Basque "populations"); a Finnish cluster (including samples from Finnish "populations") and a Spanish cluster (including samples from Spanish "populations"). Most "populations" contained individuals from clusters other than the dominant cluster for that population, there were also individuals with membership of several clusters. The results of this study are presented on a map of Europe. (Bauchet, 2007) The existence of allelic clines and the observation that the bulk of human variation is continuously distributed, has led some scientists to conclude that any categorization schema attempting to partition that variation meaningfully will necessarily create artificial truncations. (Kittles & Weiss 2003). It is for this reason, Reanne Frank argues, that attempts to allocate individuals into ancestry groupings based on genetic information have yielded varying results that are highly dependent on methodological design. Serre and Pääbo (2004) make a similar claim:
The absence of strong continental clustering in the human gene pool is of practical importance. It has recently been claimed that “the greatest genetic structure that exists in the human population occurs at the racial level” (Risch et al. 2002). Our results show that this is not the case, and we see no reason to assume that “races” represent any units of relevance for understanding human genetic history.
In a response to Serre and Pääbo (2004), Rosenberg et al. (2005) make three relevant observations. Firstly they maintain that their clustering analysis is robust. Secondly they agree with Serre and Pääbo that membership of multiple clusters can be interpreted as evidence for clinality (isolation by distance), though they also comment that this may also be due to admixture between neighbouring groups (small island model). Thirdly they comment that evidence of clusterdness is not evidence for any concepts of "biological race".
Serre and Pääbo argue that human genetic diversity consists of clines of variation in allele frequencies. We agree and had commented on this issue in our original paper: “In several populations, individuals had partial membership in multiple clusters, with similar membership coefficients for most individuals. These populations might reflect continuous gradations across regions or admixture of neighboring groups.” (Rosenberg, 2002) At the same time, we find that human genetic diversity consists not only of clines, but also of clusters, which STRUCTURE observes to be repeatable and robust....Our evidence for clustering should not be taken as evidence of our support of any particular concept of “biological race.” In general, representations of human genetic diversity are evaluated based on their ability to facilitate further research into such topics as human evolutionary history and the identification of medically important genotypes that vary in frequency across populations. Both clines and clusters are among the constructs that meet this standard of usefulness: for example, clines of allele frequency variation have proven important for inference about the genetic history of Europe, and clusters have been shown to be valuable for avoidance of the false positive associations that result from population structure in genetic association studies. The arguments about the existence or nonexistence of “biological races” in the absence of a specific context are largely orthogonal to the question of scientific utility, and they should not obscure the fact that, ultimately, the primary goals for studies of genetic variation in humans are to make inferences about human evolutionary history, human biology, and the genetic causes of disease.
Similarly Witherspoon et al. (2007) have shown that while it is possible to classify people into genetic clusters this does not resolve the observation that any two individuals from different populations are often genetically more similar to each other than to two individuals from the same population:
Discussions of genetic differences between major human populations have long been dominated by two facts: (a) Such differences account for only a small fraction of variance in allele frequencies, but nonetheless (b) multilocus statistics assign most individuals to the correct population. This is widely understood to reflect the increased discriminatory power of multilocus statistics. Yet Bamshad et al. (2004) showed, using multilocus statistics and nearly 400 polymorphic loci, that (c) pairs of individuals from different populations are often more similar than pairs from the same population. If multilocus statistics are so powerful, then how are we to understand this finding?
All three of the claims listed above appear in disputes over the significance of human population variation and "race"...The Human Genome Project (2001, p. 812) states that "two random individuals from any one group are almost as different [genetically] as any two random individuals from the entire world."
Risch et al. (2002) state that "two Caucasians are more similar to each other genetically than a Caucasian and an Asian", but Bamshad et al (2004) used the same data set as Rosenberg et al. (2002) to show that Europeans are more similar to Asians 38% of the time than they are to other Europeans when only 377 microsatellite markers are analysed.
| x | Africans | Europeans | Asians |
|---|---|---|---|
| Europeans | 36.5 | — | — |
| Asians | 35.5 | 38.3 | — |
| Indigenous Americans | 26.1 | 33.4 | 35 |
In agreement with the observation of Bamshad et al. (2004), Witherspoon et al. (2007) have shown that many more than 326 or 377 microsatellite loci are required in order to show that individuals are always more similar to individuals in their own population group than to individuals in different population groups, even for three distinct populations.
In 2007 Witherspoon et al. sought to investigate these apparently contradictory observations. In their paper Genetic similarities within and between human populations they expand upon the observation of Bamshad et al. (2004). They show that the observed clustering of human populations into relatively discrete groups is a product of using what they call "population trait values". This means that each individual is compared to the "typical" trait for several populations, and assigned to a population based on the individual's overall similarity to one of the populations as a whole: "population membership is treated as an additive quantitative genetic trait controlled by many loci of equal effect, and individuals are divided into populations on the basis of their trait values." They therefore claim that clustering analyses cannot necessarily be used to make inferences regarding the similarity or dissimilarity of individuals between or within clusters, but only for similarities or dissimilarities of individuals to the "trait values" of any given cluster. The paper measures the rate of misclassification using these "trait values" and calls this the "population trait value misclassification rate" (CT). The paper investigates the similarities between individuals by use of what they term the "dissimilarity fraction" (ω): "the probability that a pair of individuals randomly chosen from different populations is genetically more similar than an independent pair chosen from any single population." Witherspoon et al. show that two individuals can be more genetically similar to each other than to the typical genetic type of their own respective populations, and yet be correctly assigned to their respective populations. An important observation is that the likelihood that two individuals from different populations will be more similar to each other genetically than two individuals from the same population depends on several criteria, most importantly the number of genes studied and the distinctiveness of the populations under investigation.
Given 10 loci, three distinct populations, and the full spectrum of polymorphisms, the answer is ω ~ 0.3, or nearly one-third of the time. With 100 loci, the answer is ~20% of the time and even using 1000 loci, ω ~ 10%. However, if genetic similarity is measured over many thousands of loci, the answer becomes never when individuals are sampled from geographically separated populations.
By geographically separated populations, they mean sampling of people only from distant geographical regions while omitting intermediate regions, in this case Europe, sub-Saharan Africa, and East Asian. They continue:
On the other hand, if the entire world population were analyzed, the inclusion of many closely related and admixed populations would increase ω... In a similar vein, Romualdi et al. (2002) and Serre and Paabo (2004) have suggested that highly accurate classification of individuals from continuously sampled (and therefore closely related) populations may be impossible.... Classification methods typically make use of aggregate properties of populations, not just properties of individuals or even of pairs of individuals... The Structure classification algorithm (Pritchard et al. 2000) also relies on aggregate properties of populations, such as Hardy–Weinberg and linkage equilibrium. In contrast, the pairwise distances used to compute ω make no use of population-level information and are strongly affected by the high level of within-groups variation typical of human populations. This accounts for the difference in behaviour between ω and the classification results.
Witherspoon et al. also add:
given enough genetic data, individuals can be correctly assigned to their populations of origin is compatible with the observation that most human genetic variation is found within populations, not between them. It is also compatible with our finding that, even when the most distinct populations are considered and hundreds of loci are used, individuals are frequently more similar to members of other populations than to members of their own population.
Summary of different biological definitions of race
| Concept | Reference | Definition |
|---|---|---|
| Essentialist | Hooton (1926) | "A great division of mankind, characterized as a group by the sharing of a certain combination of features, which have been derived from their common descent, and constitute a vague physical background, usually more or less obscured by individual variations, and realized best in a composite picture." |
| Taxonomic | Mayr (1969) | "An aggregate of phenotypically similar populations of a species, inhabiting a geographic subdivision of the range of a species, and differing taxonomically from other populations of the species." |
| Population | Dobzhansky (1970) | "Races are genetically distinct Mendelian populations. They are neither individuals nor particular genotypes, they consist of individuals who differ genetically among themselves." |
| Lineage | Templeton (1998) | "A subspecies (race) is a distinct evolutionary lineage within a species. This definition requires that a subspecies be genetically differentiated due to barriers to genetic exchange that have persisted for long periods of time; that is, the subspecies must have historical continuity in addition to current genetic differentiation." |
Current views across disciplines
One result of debates over the meaning and validity of the concept "race" is that the current literature across different disciplines regarding human variation lacks consensus, though within some fields, such as biology, there is strong consensus. Some studies use the word race in its early essentialist taxonomic sense. Many others still use the term race, but use it to mean a population, clade, or haplogroup. Others eschew the concept of race altogether, and use the concept of population as a less problematical unit of analysis.
Since 1932, some college textbooks introducing physical anthropology have increasingly come to reject race as a valid concept: from 1932 to 1976, only seven out of thirty-two rejected race; from 1975 to 1984, thirteen out of thirty-three rejected race; from 1985 to 1993, thirteen out of nineteen rejected race. According to one academic journal entry, where 78 percent of the articles in the 1931 Journal of Physical Anthropology employed these or nearly synonymous terms reflecting a bio-race paradigm, only 36 percent did so in 1965, and just 28 percent did in 1996. The American Anthropological Association, drawing on biological research, currently holds that "The concept of race is a social and cultural construction... . Race simply cannot be tested or proven scientifically," and that, "It is clear that human populations are not unambiguous, clearly demarcated, biologically distinct groups. The concept of 'race' has no validity ... in the human species".
In an ongoing debate, some geneticists argue that race is neither a meaningful concept nor a useful heuristic device, and even that genetic differences among groups are biologically meaningless, on the grounds that more genetic variation exists within such races than among them, and that racial traits overlap without discrete boundaries. Other geneticists, in contrast, argue that categories of self-identified race/ethnicity or biogeographic ancestry are both valid and useful, that these categories correspond with clusters inferred from multilocus genetic data, and that this correspondence implies that genetic factors might contribute to unexplained phenotypic variation between groups.
In February, 2001, the editors of the medical journal Archives of Pediatrics and Adolescent Medicine asked authors to no longer use "race" as an explanatory variable and not to use obsolescent terms. Some other peer-reviewed journals, such as the New England Journal of Medicine and the American Journal of Public Health, have made similar endeavours. Furthermore, the National Institutes of Health recently issued a program announcement for grant applications through February 1, 2006, specifically seeking researchers who can investigate and publicize among primary care physicians the detrimental effects on the nation's health of the practice of medical racial profiling using such terms. The program announcement quoted the editors of one journal as saying that, "analysis by race and ethnicity has become an analytical knee-jerk reflex."
A survey, taken in 1985 (Lieberman et al. 1992), asked 1,200 American anthropologists how many disagree with the following proposition: "There are biological races in the species Homo sapiens." The responses were:
- physical anthropologists 41%
- cultural anthropologists 53%
The figure for physical anthropologists at PhD granting departments was slightly higher, rising from 41% to 42%, with 50% agreeing. This survey, however, did not specify any particular definition of race (although it did clearly specify biological race within the species Homo Sapiens); it is difficult to say whether those who supported the statement thought of race in taxonomic or population terms.
The same survey, taken in 1999, showed the following changing results for anthropologists:
- physical anthropologists 69%
- cultural anthropologists 80%
In Poland the race concept was rejected by only 25 percent of anthropologists in 2001, although: "Unlike the U.S. anthropologists, Polish anthropologists tend to regard race as a term without taxonomic value, often as a substitute for population."
In the face of these issues, some evolutionary scientists have simply abandoned the concept of race in favour of " population." What distinguishes population from previous groupings of humans by race is that it refers to a breeding population (essential to genetic calculations) and not to a biological taxon. Other evolutionary scientists have abandoned the concept of race in favour of cline (meaning, how the frequency of a trait changes along a geographic gradient). (The concepts of population and cline are not, however, mutually exclusive and both are used by many evolutionary scientists.)
According to Jonathan Marks,
- By the 1970s, it had become clear that (1)most human differences were cultural; (2) what was not cultural was principally polymorphic - that is to say, found in diverse groups of people at different frequencies; (3) what was not cultural or polymorphic was principally clinal - that is to say, gradually variable over geography; and (4) what was left - the component of human diversity that was not cultural, polymorphic, or clinal - was very small.
- A consensus consequently developed among anthropologists and geneticists that race as the previous generation had known it - as largely discrete, geographically distinct, gene pools - did not exist.
In the face of this rejection of race by evolutionary scientists, many social scientists have replaced the word race with the word "ethnicity" to refer to self-identifying groups based on beliefs concerning shared culture, ancestry and history. Alongside empirical and conceptual problems with "race," following the Second World War, evolutionary and social scientists were acutely aware of how beliefs about race had been used to justify discrimination, apartheid, slavery, and genocide. This questioning gained momentum in the 1960s during the U.S. civil rights movement and the emergence of numerous anti-colonial movements worldwide. They thus came to understood that these justifications, even when expressed in language that sought to appear objective, were social constructs.
Races as social constructions
Even as the idea of "race" was becoming a powerful organizing principle in many societies, the shortcomings of the concept were apparent. In the Old World, the gradual transition in appearances from one group to adjacent groups emphasized that "one variety of mankind does so sensibly pass into the other, that you cannot mark out the limits between them," as Blumenbach observed in his writings on human variation (Marks 1995, p. 54). As anthropologists and other evolutionary scientists have shifted away from the language of race to the term population to talk about genetic differences, Historians, anthropologists and social scientists have re-conceptualized the term "race" as a cultural category or social construct, in other words, as a particular way that some people have of talking about themselves and others. As Stephan Palmie has recently summarized, race "is not a thing but a social relation"; or, in the words of Katya Gibel Mevorach, "a metonym," "a human invention whose criteria for differentiation are neither universal nor fixed but have always been used to manage difference." As such it cannot be a useful analytical concept; rather, the use of the term "race" itself must be analyzed. Moreover, they argue that biology will not explain why or how people use the idea of race: history and social relationships will. For example, the fact that in many parts of the United States, categories such as Hispanic or Latino are viewed to constitute a race (instead of an ethnic group) reflect this new idea of "race as a social construct". However, it may be in the interest of dominant groups to cluster Spanish speakers into a single, isolated population, rather than classifying them according to Race (as are the rest of U.S. racial groups). Especially in the context of the debate over immigration. "According to the 2000 census, two-thirds [of Hispanics] are of Mexican heritage . . . So, for practical purposes, when we speak of Hispanics and Latinos in the U.S., we’re really talking about Native Americans . . . [therefore] if being Hispanic carries any societal consequences that justify inclusion in the pantheon of great American racial minorities, they’re the result of having Native American blood. [But imagine the] the impact this would have on the illegal-immigration debate. It’s one thing to blame the fall of western civilization on illegal Mexican immigration, but quite thornier to blame it on illegal Amerindian immigration from Mexico."
In the United States
The immigrants to the New World came largely from widely separated regions of the Old World—western and northern Europe, West Africa, and, later, eastern Asia and southern Europe. In the Americas, the immigrant populations began to mix among themselves and with the indigenous inhabitants of the continent. In the United States, for example, most people who self-identify as African American have some European ancestors — in one analysis of genetic markers that have differing frequencies between continents, European ancestry ranged from an estimated 7% for a sample of Jamaicans to ∼23% for a sample of African Americans from New Orleans (Parra et al. 1998). Similarly, many people who identify as European American have some African or Native American ancestors, either through openly interracial marriages or through the gradual inclusion of people with mixed ancestry into the majority population. In a survey of college students who self-identified as white in a northeastern U.S. university, ∼30% were estimated to have less than 90% European ancestry.
In the United States since its early history, Native Americans, African-Americans and European-Americans were classified as belonging to different races. For nearly three centuries, the criteria for membership in these groups were similar, comprising a person’s appearance, his fraction of known non-White ancestry, and his social circle.2 But the criteria for membership in these races diverged in the late 19th century. During Reconstruction, increasing numbers of Americans began to consider anyone with " one drop" of known "Black blood" to be Black regardless of appearance.3 By the early 20th century, this notion of invisible blackness was made statutory in many states and widely adopted nationwide.4 In contrast, Amerindians continue to be defined by a certain percentage of "Indian blood" (called blood quantum) due in large part to American slavery ethics. Finally, for the past century or so, to be White one had to have perceived "pure" White ancestry.
Efforts to sort the increasingly mixed population of the United States into discrete categories generated many difficulties (Spickard 1992). By the standards used in past censuses, many millions of children born in the United States have belonged to a different race than have one of their biological parents. Efforts to track mixing between groups led to a proliferation of categories (such as "mulatto" and "octoroon") and "blood quantum" distinctions that became increasingly untethered from self-reported ancestry. A person's racial identity can change over time, and self-ascribed race can differ from assigned race (Kressin et al. 2003). Until the 2000 census, Latinos were required to identify with a single race despite the long history of mixing in Latin America; partly as a result of the confusion generated by the distinction, 32.9% (U.S. census records) of Latino respondents in the 2000 census ignored the specified racial categories and checked "some other race". (Mays et al. 2003 claim a figure of 42%)
The difference between how Native American and Black identities are defined today (blood quantum versus one-drop) has demanded explanation. According to anthropologists such as Gerald Sider, the goal of such racial designations was to concentrate power, wealth, privilege and land in the hands of Whites in a society of White hegemony and privilege (Sider 1996; see also Fields 1990). The differences have little to do with biology and far more to do with the history of racism and specific forms of White supremacy (the social, geopolitical and economic agendas of dominant Whites vis-à-vis subordinate Blacks and Native Americans) especially the different roles Blacks and Amerindians occupied in White-dominated 19th century America. The theory suggests that the blood quantum definition of Native American identity enabled Whites to acquire Amerindian lands, while the one-drop rule of Black identity enabled Whites to preserve their agricultural labor force. The contrast presumably emerged because as peoples transported far from their land and kinship ties on another continent, Black labor was relatively easy to control, thus reducing Blacks to valuable commodities as agricultural laborers. In contrast, Amerindian labor was more difficult to control; moreover, Amerindians occupied large territories that became valuable as agricultural lands, especially with the invention of new technologies such as railroads; thus, the blood quantum definition enhanced White acquisition of Amerindian lands in a doctrine of Manifest Destiny that subjected them to marginalization and multiple episodic localized campaigns of extermination.
The political economy of race had different consequences for the descendants of aboriginal Americans and African slaves. The 19th century blood quantum rule meant that it was relatively easier for a person of mixed Euro-Amerindian ancestry to be accepted as White. The offspring of only a few generations of intermarriage between Amerindians and Whites likely would not have been considered Amerindian at all (at least not in a legal sense). Amerindians could have treaty rights to land, but because an individual with one Amerindian great-grandparent no longer was classified as Amerindian, they lost any legal claim to Amerindian land. According to the theory, this enabled Whites to acquire Amerindian lands. The irony is that the same individuals who could be denied legal standing because they were "too White" to claim property rights, might still be Amerindian enough to be considered as " breeds", stigmatized for their Native American ancestry.
The 20th century one-drop rule, on the other hand, made it relatively difficult for anyone of known Black ancestry to be accepted as White. The child of a Black sharecropper and a White person was considered Black. And, significant in terms of the economics of sharecropping, such a person also would likely be a sharecropper as well, thus adding to the employer's labor force.
In short, this theory suggests that in a 20th century economy that benefited from sharecropping, it was useful to have as many Blacks as possible. Conversely, in a 19th century nation bent on westward expansion, it was advantageous to diminish the numbers of those who could claim title to Amerindian lands by simply defining them out of existence.
It must be mentioned, however, that although some scholars of the Jim Crow period agree that the 20th century notion of invisible Blackness shifted the color line in the direction of paleness, thereby swelling the labor force in response to Southern Blacks' great migration northwards, others (Joel Williamson, C. Vann Woodward, George M. Fredrickson, Stetson Kennedy) see the one-drop rule as a simple consequence of the need to define Whiteness as being pure, thus justifying White-on-Black oppression. In any event, over the centuries when Whites wielded power over both Blacks and Amerindians and widely believed in their inherent superiority over people of colour, it is no coincidence that the hardest racial group in which to prove membership was the White one.
In the United States, social and legal conventions developed over time that forced individuals of mixed ancestry into simplified racial categories (Gossett 1997). An example is the " one-drop rule" implemented in some state laws that treated anyone with a single known African American ancestor as black (Davis 2001). The decennial censuses conducted since 1790 in the United States also created an incentive to establish racial categories and fit people into those categories (Nobles 2000). In other countries in the Americas where mixing among groups was overtly more extensive, social categories have tended to be more numerous and fluid, with people moving into or out of categories on the basis of a combination of socioeconomic status, social class, ancestry, and appearance (Mörner 1967).
The term " Hispanic" as an ethnonym emerged in the 20th century with the rise of migration of laborers from American Spanish-speaking countries to the United States. It includes people who had been considered racially distinct (Black, White, Amerindian, Asian, and mixed groups) in their home countries. Today, the word "Latino" is often used as a synonym for "Hispanic". In contrast to "Latino"´or "Hispanic" " Anglo" is now used to refer to non-Hispanic White Americans or non-Hispanic European Americans, most of whom speak the English language but are not necessarily of English descent.
In Brazil
Compared to 19th century United States, 20th century Brazil was characterized by a perceived relative absence of sharply defined racial groups. According to anthropologist Marvin Harris (1989), this pattern reflects a different history and different social relations. Basically, race in Brazil was "biologized," but in a way that recognized the difference between ancestry (which determines genotype) and phenotypic differences. There, racial identity was not governed by such a rigid descent rule as in the United States. A Brazilian child was never automatically identified with the racial type of one or both parents, nor were there only a very limited number of categories to choose from. Over a dozen racial categories would be recognized in conformity with all the possible combinations of hair color, hair texture, eye color, and skin colour. These types grade into each other like the colors of the spectrum, and no one category stands significantly isolated from the rest. That is, race referred preferencially to appearance, not heredity.
Through this system of racial identification, parents and children and even brothers and sisters were frequently accepted as representatives of completely different racial types. In a fishing village in the state of Bahia, an investigator showed 100 people pictures of three sisters and asked them to identify the races of each. In only six responses were the sisters identified by the same racial term. Fourteen responses used a different term for each sister (Harris 1964: 57). In another experiment nine portraits were shown to a hundred people. Forty different racial types were elicited (Harris 1964: 58). It was found, in addition, that a given Brazilian might be called by as many as thirteen different terms by other members of the community (Harris 1964: 57). These terms are spread out across practically the entire spectrum of theoretical racial types. A further consequence of the absence of a descent rule was that Brazilians apparently not only disagreed about the racial identity of specific individuals, but they also seemed to be in disagreement about the abstract meaning of the racial terms as defined by words and phrases. For example, 40% of a sample ranked moreno claro ("light" person of primarily European ancestry with dark hair) as a lighter type than mulato claro ("light" person of mixed European and African ancestry), while 60% reversed this order (Harris 1964: 58). A further note of confusion is that one person might employ different racial terms to describe the same person over a short time span (Harris 1964: 59; Goldstein 1999: 566-568).} [For a solid discussion of Brazilian racial terms, see Livio Sansone's Blackness Without Ethnicity (2003) and France Winddance Twine's Racism in a Racial Democracy (1998).] The choice of which racial description to use may vary according to the relationship (be it personal, class-based, or otherwise) between the speaker and the person concerned and moods of the individuals involved (Harris 1964: 59).
So, although the identification of a person by race is far more fluid and flexible in Brazil than in the U.S., there still are racial stereotypes and prejudices. African features have been considered less desirable; Blacks have been considered socially inferior, and Whites superior (Harris 1964: 59-60). These white supremacist values seem to be an obvious legacy of Portuguese colonization and the slave-based plantation system (Harris 1964: 54-57). The complexity of racial classifications in Brazil is reflective of the extent of miscegenation in Brazilian society, a society that remains highly, but not strictly, stratified along colour lines. Henceforth, the Brazilian narrative of a perfect "post-racist" country, must be met with caution, as sociologist Gilberto Freyre demonstrated in 1933 in Casa Grande e Senzala.
Marketing of race: genetic lineages as social lineages
New research in molecular genetics, and the marketing of genetic identities through the analysis of one's Y chromosome, mtDNA or autosomal DNA, has reignited the debate surrounding race. Most of the controversy surrounds the question of how to interpret these new data, and whether conclusions based on existing data are sound. Although the vast majority of researchers endorse the view that continental groups do not constitute different subspecies, and molecular geneticists generally reject the identification of mtDNA and Y chromosomal lineages or allele clusters with "races", some anthropologists have suggested that the marketing of genetic analysis to the general public in the form of "Personalized Genetic Histories" (PGH) is leading to a new social construction of race. See above sections Molecular lineages, Y chromosomes and mitochondrial DNA and How much are genes shared? Clustering analyses and what they tell us.
Typically, a consumer of a commercial PGH service sends in a sample of DNA which is analyzed by molecular biologists and is sent a report, of which the following is a sample
- "African DNA Ancestry Report"
The subject's likely haplogroup L2 is associated with the so-called Bantu expansion from West and Central sub-Saharan Africa east and south, dated 2,000-4,000 years ago .... Between the 15th and 19th centuries C.E, the Atlantic slave trade resulted in the forced movement of approximately 13 million people from Africa, mainly to the Americas. Only approximately 11 million survived the passage and many more died in the early years of captivity. Many of these slaves were traded to the West African Cape Verde ports of embarkation through Portuguese and Arab middlemen and came from as far south as Angola. Among the African tribal groups, all Bantu-speaking, in which L2 is common are: Hausa, Kanuri, Fulfe, Songhai, Malunjin (Angola), Yoruba, Senegalese, Serer and Wolof.
Although no single sentence in such a report is technically wrong, through the combination of these sentences, anthropologists and others have argued, the report is telling a story that connects a haplotype with a language and a group of tribes. This story is generally rejected by research scientists for the simple reason that an individual receives his or her Y chromosome or mtDNA from only one ancestor in every generation; consequently, with every generation one goes back in time, the percentage of ones ancestors it represents halves; if one goes back hundreds (let alone thousands) of years, it represents only a tiny fragment of one's ancestry. As Mark Shriver and Rick Kittles recently remarked,
For many customers of lineage-based tests, there is a lack of understanding that their maternal and paternal lineages do not necessarily represent their entire genetic make-up. For example, an individual might have more than 85% Western European 'genomic' ancestry but still have a West African mtDNA or NRY lineage.
Nevertheless, they acknowledge, such stories are increasingly appealing to the general public. Thus, in his book Blood of the Isles (published in the US and Canada as Saxons, Vikings and Celts: The Genetic Roots of Britain and Ireland), however, Bryan Sykes discusses how people who have been mtDNA tested by his commercial laboratory and been found to belong to the same haplogroup have parties together because they see this as some sort of "bond", even thought these people may not actually share very much ancestry.
Through these kinds of reports, new advances in molecular genetics are being used to create or confirm stories have about social identities. Although these identities are not racial in the biological sense, they are in the cultural sense in that they link biological and cultural identities. Nadia Abu el-Haj has argued that the significance of gentetic lineages in popular conceptions of race owes to the perception that while genetic lineages, like older notions of race, suggests some idea of biological relatedness, unlike older notions of race they are not directly connected to claims about human behaviour or character. Abu el-Haj has thus argued that "postgenomics does seem to be giving race a new lease on life." Nevertheless, Abu el-Haj argues that in order to understand what it means to think of race in terms of genetic lineages or clusters, one must understand that
Race science was never just about classification. It presupposed a distinctive relationship between "nature" and "culture," understanding the differences in the former to ground and to generate the different kinds of persons ("natural kinds") and the distinctive stages of cultures and civilizations that inhabit the world.
Abu el-Haj argues that genomics and the mapping of lineages and clusters liberates "the new racial science from the older one by disentangling ancestry from culture and capacity." As an example, she refers to recent work by Hammer et al., which aimed to test the claim that present-day Jews are more closely related to one another than to neighbouring non-Jewish populations. Hammer et. al found that the degree of genetic similarity among Jews shifted depending on the locus investigated, and suggested that this was the result of natural selection acting on particular loci. They therefore focused on the non-recombining Y chromosome to "circumvent some of the complications associated with selection". As another example she points to work by Thomas et al., who sought to distinguish between the Y chromosomes of Jewish priests (in Judaism, membership in the priesthood is passed on through the father's line) and the Y chromosomes of non-Jews. Abu el-Haj concluded that this new "race science" calls attention to the importance of "ancestry" (narrowly defined, as it does not include all ancestors) in some religions and in popular culture, and peoples' desire to use science to confirm their claims about ancestry; this "race science," she argues is fundamentally different from older notions of race that were used to explain differences in human behaviour or social status:
As neutral markers, junk DNA cannot generate cultural, behavioural, or, for that matter, truly biological differences between groups .... mtDNA and Y-chromosome markers relied on in such work are not "traits" or "qualities" in the old racial sense. They do not render some populations more prone to violence, more likely to suffer psychiatric disorders, or for that matter, incapable of being fully integrated - because of their lower evolutionary development - into a European cultural world. Instead, they are "marks," signs of religious beliefs and practices .... it is via biological noncoding genetic evidence that one can demonstrate that history itself is shared, that historical traditions are (or might well be) true."
On the other hand, there are tests that do not rely on molecular lineages, but rather on correlations between allele frequencies, often when allele frequencies correlate these are called clusters. Clustering analyses are less powerful than lineages because they cannot tell an historical story, they can only estimate the proportion of a person's ancestry from any given large geographical region. These sorts of tests use informative alleles called Ancestry-informative marker (AIM), which although shared across all human populations vary a great deal in frequency between groups of people living in geographically distant parts of the world. These tests use contemporary people sampled from certain parts of the world as references to determine the likely proportion of ancestry for any given individual. In a recent Public Service Broadcasting (PBS) programme on the subject of genetic ancestry testing the academic Henry Louis Gates: "wasn’t thrilled with the results (it turns out that 50 percent of his ancestors are likely European)". Charles Rotimi, of Howard University's National Human Genome Centre, is one of many who have highlighted the methodological flaws in such research - that "the nature or appearance of genetic clustering (grouping) of people is a function of how populations are sampled, of how criteria for boundaries between clusters are set, and of the level of resolution used" all bias the results - and concluded that people should be very cautious about relating genetic lineages or clusters to their own sense of identity. (see also above section How much are genes shared? Clustering analyses and what they tell us)
Thus, in analyses that assign individuals to groups it becomes less apparent that self-described racial groups are reliable indicators of ancestry. One cause of the reduced power of the assignment of individuals to groups is admixture. For example, self-described African Americans tend to have a mix of West African and European ancestry. Shriver et al. (2003) found that on average African Americans have ~80% African ancestry. Also, in a survey of college students who self-identified as “white” in a northeastern U.S. university, ~30% of whites had less than 90% European ancestry.
Stephan Palmie has responded to Abu el-Haj's claim that genetic lineages make possible a new, politically, economically, and socially benign notion of race and racial difference by suggesting that efforts to link genetic history and personal identity will inevitably "ground present social arrangements in a time-hallowed past," that is, use biology to explain cultural differences and social inequalities.
Political and practical uses
Racism
Race and intelligence
Researchers have reported differences in the average IQ test scores of various ethnic groups. The interpretation, causes, accuracy and reliability of these differences are highly controversial. Some researchers, such as Arthur Jensen, Richard Herrnstein, and Richard Lynn have argued that such differences are at least partially genetic. Others, for example Thomas Sowell, argue that the differences largely owe to social and economic inequalities. Still others have such as Stephen Jay Gould and Richard Lewontin have argued that categories such as "race" and "intelligence" are cultural constructs that render any attempt to explain such differences (whether genetically or sociologically) meaningless.
The Flynn effect is the rise of average Intelligence Quotient (IQ) test scores, an effect seen in most parts of the world, although at varying rates. Scholars therefore believe that rapid increases in average IQ seen in many places are much too fast to be as a result of changes in brain physiology and more likely as a result of environmental changes. The fact that environment has a significant effect on IQ demolishes the case for the use of IQ data as a source of genetic information.
In biomedicine
There is an active debate among biomedical researchers about the meaning and importance of race in their research. The primary impetus for considering race in biomedical research is the possibility of improving the prevention and treatment of diseases by predicting hard-to-ascertain factors on the basis of more easily ascertained characteristics. Some have argued that in the absence of cheap and widespread genetic tests, racial identification is the best way to predict for certain diseases, such as Cystic fibrosis, Lactose intolerance, Tay-Sachs Disease and sickle cell anaemia, which are genetically linked and more prevalent in some populations than others. The most well-known examples of genetically-determined disorders that vary in incidence among populations would be sickle cell disease, thalassaemia, and Tay-Sachs disease.
There has been criticism of associating disorders with race. For example, in the United States sickle cell is typically associated with black people, but this trait is also found in people of Mediterranean, Middle Eastern or Indian ancestry. The sickle cell trait offers some resistance to malaria. In regions where malaria is present sickle cell has been positively selected and consequently the proportion of people with it is greater. Therefore, it has been argued that sickle cell should not be associated with a particular race, but rather with having ancestors who lived in a malaria-prone region. Africans living in areas where there is no malaria, such as the East African highlands, have prevalence of sickle cell as low as parts of Northern Europe.
Another example of the use of race in medicine is the recent U.S. FDA approval of BiDil, a medication for congestive heart failure targeted at black people in the United States. Several researchers have questioned the scientific basis for arguing the merits of a medication based on race, however. As Stephan Palmie has recently pointed out, black Americans were disproportionately affected by Hurricane Katrina, but for social and not climatological reasons; similarly, certain diseases may disproportionately affect different races, but not for biological reasons. Several researchers have suggested that BiDil was re-designated as a medicine for a race-specific illness because its manufacturer, Nitromed, needed to propose a new use for an existing medication in order to justify an extension of its patent and thus monopoly on the medication, not for pharmacological reasons.
Gene flow and intermixture also have an effect on predicting a relationship between race and "race linked disorders". Multiple sclerosis is typically associated with people of European descent and is of low risk to people of African descent. However, due to gene flow between the populations, African Americans have elevated levels of MS relative to Africans. Notable African Americans affected by MS include Richard Pryor and Montel Williams. As populations continue to mix, the role of socially constructed races may diminish in identifying diseases.
In law enforcement

In an attempt to provide general descriptions that may facilitate the job of law enforcement officers seeking to apprehend suspects, the United States FBI employs the term "race" to summarize the general appearance (skin colour, hair texture, eye shape, and other such easily noticed characteristics) of individuals whom they are attempting to apprehend. From the perspective of law enforcement officers, it is generally more important to arrive at a description that will readily suggest the general appearance of an individual than to make a scientifically valid categorization by DNA or other such means. Thus in addition to assigning a wanted individual to a racial category, such a description will include: height, weight, eye colour, scars and other distinguishing characteristics, etc. Scotland Yard use a classification based in the ethnic background of British society: W1 (White-British), W2 (White-Irish), W9 (Any other white background); M1 (White and black Caribbean), M2 (White and black African), M3 (White and Asian), M9 (Any other mixed background); A1 (Asian-Indian), A2 (Asian-Pakistani), A3 (Asian-Bangladeshi), A9 (Any other Asian background); B1 (Black Caribbean), B2 (Black African), B3 (Any other black background); O1 (Chinese), O9 (Any other). Some of the characteristics that constitute these groupings are biological and some are learned (cultural, linguistic, etc.) traits that are easy to notice.
In many countries, such as France, the state is legally banned from maintaining data based on race, which often makes the police issue wanted notices to the public that include labels like "dark skin complexion", etc. One of the factors that encourages this kind of circuitous wordings is that there is controversy over the actual relationship between crimes, their assigned punishments, and the division of people into the so called "races," leading officials to try to deemphasize the alleged race of suspects. In the United States, the practice of racial profiling has been ruled to be both unconstitutional and also to constitute a violation of civil rights. There is active debate regarding the cause of a marked correlation between the recorded crimes, punishments meted out, and the country's "racially divided" people. Many consider de facto racial profiling an example of institutional racism in law enforcement. The history of misuse of racial categories to adversely impact one or more groups and/or to offer protection and advantage to another has a clear impact on debate of the legitimate use of known phenotypical or genotypical characteristics tied to the presumed race of both victims and perpetrators by the government.
More recent work in racial taxonomy based on DNA cluster analysis (see Lewontin's Fallacy) has led law enforcement to narrow their search for individuals based on a range of phenotypical characteristics found consistent with DNA evidence.
While controversial, DNA analysis has been successful in helping police identify both victims and perpetrators by giving an indication of what phenotypical characteristics to look for and what community the individual may have lived in. For example, in one case phenotypical characteristics suggested that the friends and family of an unidentified victim would be found among the Asian community, but the DNA evidence directed official attention to missing Native Americans, where her true identity was eventually confirmed. In an attempt to avoid potentially misleading associations suggested by the word "race," this classification is called "biogeographical ancestry" (BGA), but the terms for the BGA categories are similar to those used as for race. The difference is that ancestry-informative DNA markers identify continent-of-ancestry admixture, not ethnic self-identity, and provide a wide range of phenotypical characteristics such that some people in a biogeographical category will not match the stereotypical image of an individual belonging to the corresponding race. To facilitate the work of officials trying to find individuals based on the evidence of their DNA traces, firms providing the genetic analyses also provide photographs showing a full range of phenotypical characteristics of people in each biogeographical group. Of special interest to officials trying to find individuals on the basis of DNA samples that indicate a diverse genetic background is what range of phenotypical characteristics people with that general mixture of genotypical characteristics may display.
Similarly, forensic anthropologists draw on highly heritable morphological features of human remains (e.g. cranial measurements) in order to aid in the identification of the body, including in terms of race. In a recent article anthropologist Norman Sauer asked, "if races don't exist, why are forensic anthropologists so good at identifying them." Sauer observed that the use of 19th century racial categories is widespread among forensic anthropologists:
- "In many cases there is little doubt that an individual belonged to the Negro, Caucasian, or Mongoloid racial stock."
- "Thus the forensic anthropologist uses the term race in the very broad sense to differentiate what are commonly known as white, black and yellow racial stocks."
- "In estimating race forensically, we prefer to determine if the skeleton is Negroid, or Non-Negroid. If findings favour Non-Negroid, then further study is necessary to rule out Mongoloid."
According to Sauer, "The assessment of these categories is based upon copious amounts of research on the relationship between biological characteristics of the living and their skeletons." Nevertheless, he agrees with other anthropologists that race is not a valid biological taxonomic category, and that races are socially constructed. He argued there is nevertheless a strong relationship between the phenotypic features forensic anthropologists base their identifications on, and popular racial categories. Thus, he argued, forensic anthropologists apply a racial label to human remains because their analysis of physical morphology enables them to predict that when the person was alive, that particular racial label would have been applied to them.